Analysis and Enrichment of Finnish Unstructured Text Data
Some Background¶
In my personal experiences with natural language processing (NLP) projects, Finnish language's agglutinative nature adds another dimension of difficulty in comparison to Germanic languages. Since Finno-Ugrian languages are quite deviated in their grammar from Germanic languages, there are less sophisticated and easy to use tools available for Finnish NLP. However, there still are some tools available, such as TurkuNLP, FiNER and uralicNLP for morphological analysis and tagging, though setting up these tools can be tricky for a newbie such as myself.
I'm quite interested in named entity tagging (NER), since it can be used for text pseudonymization, which can be really valuable for electronic health records (EHRs). EHRs contain many types of information, but I'm interested in the free text type of information, since so much of it remain underutilized and it holds immense amounts of information. However, EHRs contain sensitive information about private individuals, such as names, addresses and so on. Therefore, to protect individuals privacy, non-pseudonymized EHRs shouldn't be given out to researchers (if you want to know more, check out my previous blog about EHRs).
If the text can be pseudonymized, it can be more accessible for research as individual's information security remains unthreatened. But if we can automate recognization of named entities, couldn't that also be used for text enrichment? With text enrichment, one can add another dimension of useful information into text data, that will be demonstrated in following sections. Therefore I decided to bite the bullet and try out named entity tagging with FiNER on Finnish text dataset.
Collecting and Inspecting the Dataset¶
I used Kielipankki's Korp-corpus builder service. I chose the Suomi24 2001–2017 forum discussions as base dataset from which I queried messages with the word "science" (fin. tiede) in them. I chose first 3000 entries as my base dataset. It could be useful to inspect the Korp dataset before named entity tagging with FiNER. Superficial analysis, such as word counts and word pair counts, can be useful to characterize my dataset and understand what kind of words are enriched in the dataset. It is not necessary to stem or lemmatize words in the dataset, since Korp has readily lemmatized the words. Lemmatization means turning the word inflections into their base forms. This makes handling the text data much easier.
import nltk
from nltk.tokenize import sent_tokenize, word_tokenize
from nltk.corpus import stopwords
import pandas as pd
# import the data, I downloaded the dataset as three datasets of thousand messages each
# there were high amount of duplicates in the third .tsv file, so I cut off the last 200 rows
df1 = pd.read_csv("KorpData/korp_kwic_tiede_tiede_1.tsv", sep="\t", engine='python', encoding="utf-8")
df2 = pd.read_csv("KorpData/korp_kwic_tiede_tiede_2.tsv", sep="\t", engine='python', encoding="utf-8")
df3 = pd.read_csv("KorpData/korp_kwic_tiede_tiede_3.tsv", sep="\t", engine='python', encoding="utf-8").iloc[0:800, :]
testidata = pd.DataFrame()
testidata = testidata.append([df1, df2, df3])
# I'll import Finnish stopwords, which include usually frequent, but semantically meaningless words
stopWords = set(stopwords.words('finnish'))
# Here I define special characters that I'll filter from the dataset along with stopwords
specials = [",", ".", ">", "<", ";", ":", "!", "?", "[", "]", "{", "}", "(", ")", "/", "="]
wordsFiltered = []
stemmedWords = []
tokens = []
# here I split "lemmas" column's cells entries into individual words
for teksti in testidata['lemmas']:
tokens.append(word_tokenize(teksti))
# here I'll concatenate each list of words into one single list of words
tokens = list(itertools.chain.from_iterable(tokens))
# filter words if they are stopwords or special characters
for token in tokens:
if token not in stopWords and token not in specials:
wordsFiltered.append(token)
wordfreq = {}
# here I actually calculate the frequency of unique words and save counts into a dictionary
for word in wordsFiltered: #
if word not in wordfreq.keys():
wordfreq[word] = 1
else:
wordfreq[word] += 1
# filter words for word pair counts
wordsFiltered2 = []
for word in wordsFiltered:
if len(word) > 2: # Here I include words that are over 3 characters in length
wordsFiltered2.append(word)
else:
pass
bigrams = (list(nltk.bigrams(wordsFiltered2))) # here I create the wordpairs
wordfreq2 = {}
# count the frequency of unique wordpairs and save them into a dictionary
for bigram in bigrams:
if bigram not in wordfreq2.keys():
wordfreq2[bigram] = 1
else:
wordfreq2[bigram] += 1
Plot the Word Counts:¶
Here I'll inspect what are the most enriched words in the dataset. I simply create a dataframe from word count dictionary and seaborn will do the rest.
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
# create the dataframe
df = pd.DataFrame.from_dict(wordfreq, orient='index', columns=['frekvenssi'])
df = df.reset_index()
# filter words that are three characters or less in length
df2 = df[df['index'].map(len) > 3]
# plot the frequencies
plt.figure(figsize=[9,10], dpi=80)
sns.set_style("darkgrid")
plt.title("korp avainsana: tiede, sanojen frekvenssit")
sns.barplot(x="frekvenssi", y="index", data=df2.sort_values(by='frekvenssi', ascending=False).head(40))
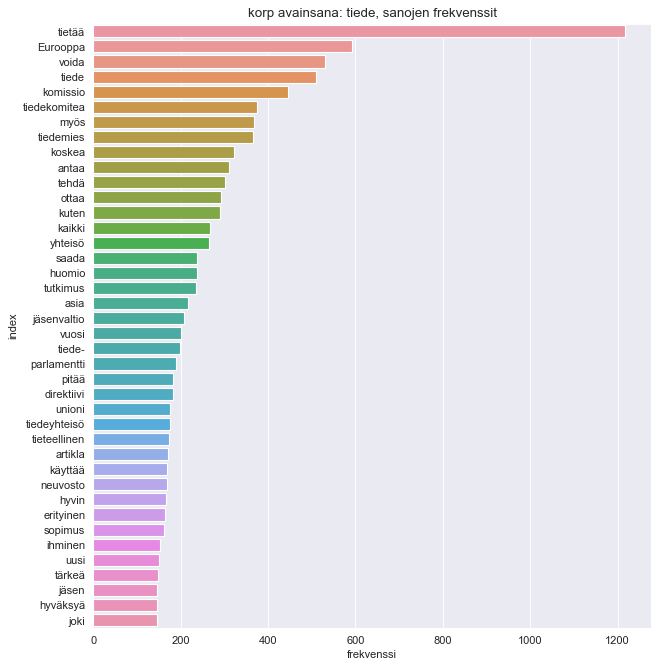
Now that's interesting, I didn't take into consideration that the word tiede also has inflections of tiedetään (we know/is known), tiedetty (has been known), or tiedossa (is known/noted) to name a few, which in their lemmatized forms turn into form tietää (knows). Also certain bureaucratic terms are quite frequent in the dataset.
Plot the Word Pair Counts:¶
Process will essentially be the same as for word counts.
dfbg = pd.DataFrame.from_dict(wordfreq2, orient='index', columns=['frekvenssi']) # create the datagrame
dfbg = dfbg.reset_index()
plt.figure(figsize=[9,10], dpi=80) # plot word pair counts
sns.set_style("darkgrid")
plt.title("korp avainsana: tiede, bigramien frekvenssit")
sns.barplot(x="frekvenssi", y="index", data=dfbg.sort_values(by='frekvenssi', ascending=False).head(40))
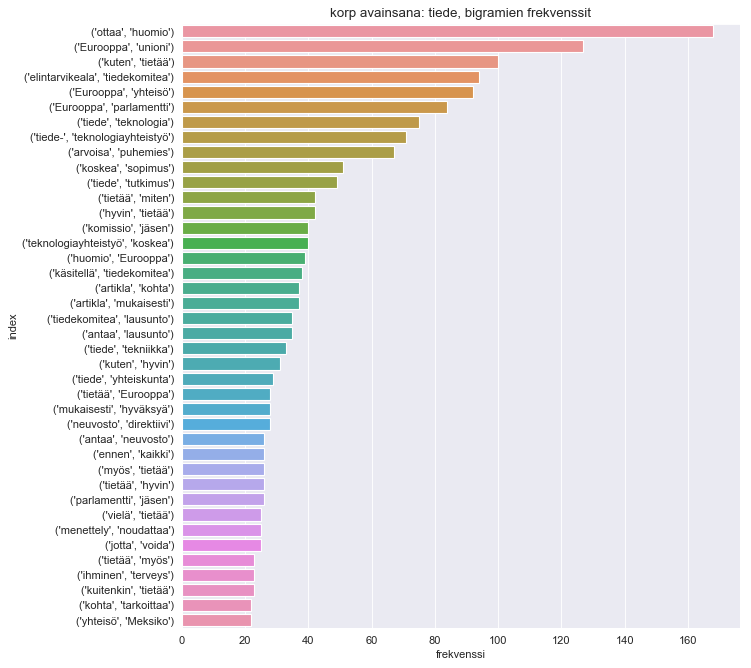
There are certainly quite generic bigrams present, such as ottaa huomio (take note) or Eurooppa unioni (European union). However, these bigrams can explain what kind of context are most frequent words talked in.
Word Correlation Analysis¶
While bigram frequencies can be useful for finding word pair enrichments, word correlation could be useful in discovering words that co-occur but that are not right next to each other.
import numpy as np
# first, I'll create a bag of words (bow) dataframe
# in bow, each word in word counts dataframe (df2) will be a column in bag of words dataframe
# we'll choose words that have frequency of more than 100
bow = pd.DataFrame(0, index=np.arange(len(testidata)), columns = list(list(df2["index"][df2["frekvenssi"] > 100])))
# counting the frequency of each word in each forum message
for i in range(len(testidata["lemmas"])):
text = word_tokenize(list(testidata["lemmas"])[i])
for word in text:
for column in bow.columns:
if str(word) == str(column):
bow.loc[i, str(column)] += 1
# what is handy about pandas is that it has integrated correlation analysis:
# I'll want to inspect just the linear correlation, so I'll use Pearson method
corrPears = bow.corr(method='pearson')
#corrKend = bow.corr(method='kendall')
#corrSpear = bow.corr(method='spearman')
# then I'll print word correlations that have correlation value above 0.5 or below -0.5
# I didn't include correlation value of 1.0 because that'll just include same word pairs (e.g. komissio komissio 1.0)
print("\n---PEARSON WORD CORRELATIONS---")
indexList = []
for column in corrPears.columns:
for index in corrPears.index:
if (corrPears.loc[index, column] > 0.5 or corrPears.loc[index, column] < -0.5) and corrPears.loc[index, column] < 1.0:
if [column, index] not in indexList: # duplicate check, ensures that word pairs are not presented twice
indexList.append([index, column])
print(index, column, str(round(corrPears.loc[index, column], 3)))
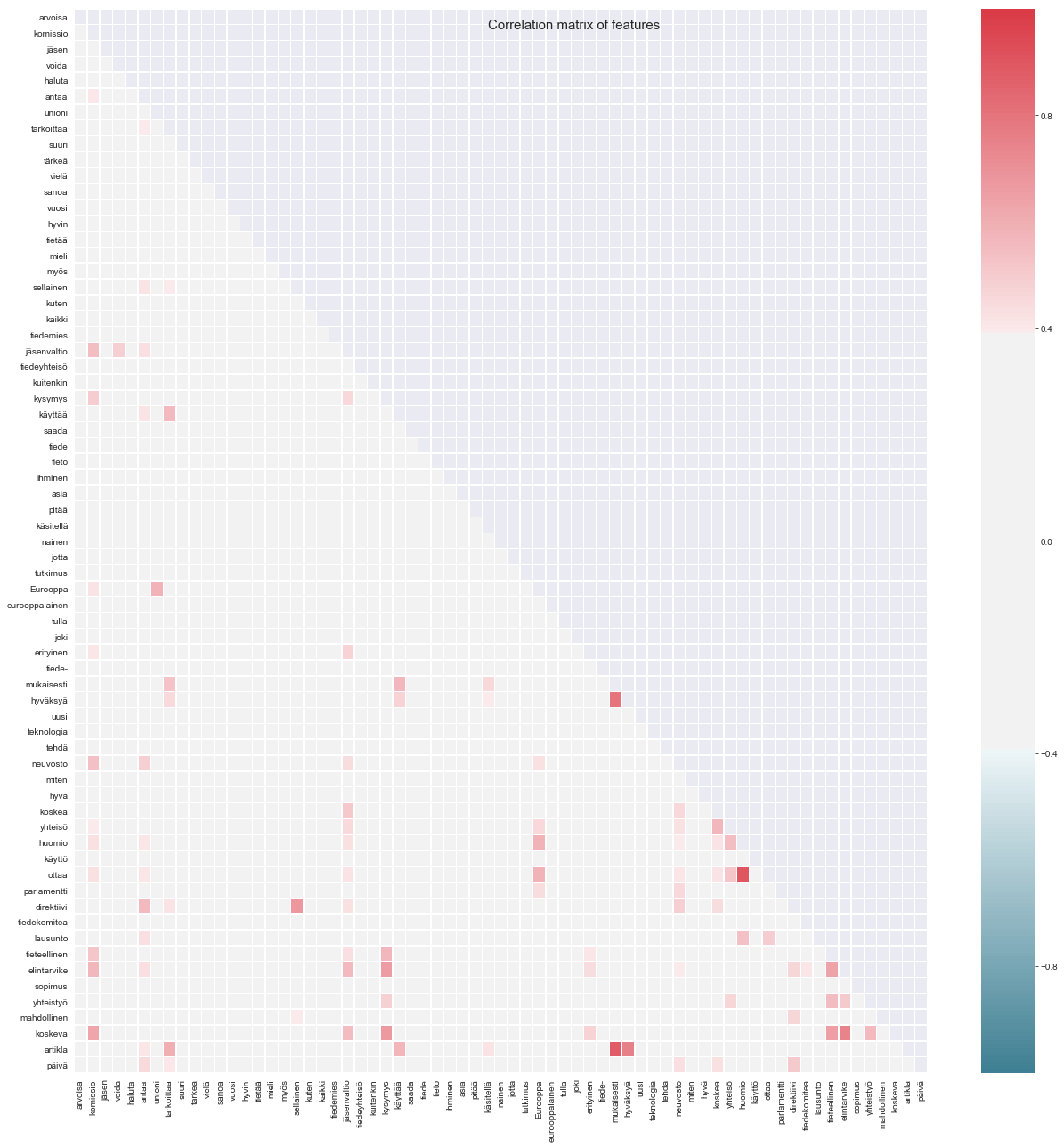
---PEARSON WORD CORRELATIONS--- - jäsenvaltio komissio 0.543 - neuvosto komissio 0.536 - tieteellinen komissio 0.517 - elintarvike komissio 0.568 - koskeva komissio 0.629 - direktiivi antaa 0.559 - Eurooppa unioni 0.582 - käyttää tarkoittaa 0.556 - mukaisesti tarkoittaa 0.524 - artikla tarkoittaa 0.597 - direktiivi sellainen 0.676 - koskea jäsenvaltio 0.509 - elintarvike jäsenvaltio 0.558 - koskeva jäsenvaltio 0.549 - tieteellinen kysymys 0.567 - elintarvike kysymys 0.658 - koskeva kysymys 0.668 - mukaisesti käyttää 0.565 - artikla käyttää 0.576 - huomio Eurooppa 0.585 - ottaa Eurooppa 0.581 - hyväksyä mukaisesti 0.8 - artikla mukaisesti 0.882 - artikla hyväksyä 0.755 - yhteisö koskea 0.564 - huomio yhteisö 0.541 - ottaa yhteisö 0.521 - ottaa huomio 0.9 - lausunto huomio 0.538 - päivä direktiivi 0.507 - elintarvike tieteellinen 0.639 - yhteistyö tieteellinen 0.549 - koskeva tieteellinen 0.65 - yhteistyö elintarvike 0.504 - koskeva elintarvike 0.75 - koskeva yhteistyö 0.556
Interesting, two semantically linked word pairs: ottaa huomio (take into consideration) and artikla mukaisesti (according to article) have quite strong correlations of 0.9 and 0.88. Others such as koskeva elintarvike (particular food product) and artikla hyväksyä (article accept) have also somewhat strong correlations ~0.75, and they are not present in top 40 most frequent bigrams.
Let's see what these correlations look like in a heatmap:
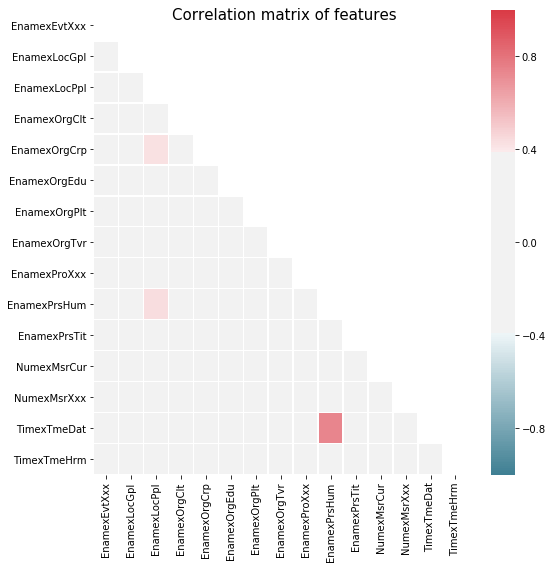
Named Entity Tagging with FiNER¶
Setting Up the Dataset for FiNER¶
FiNER processes individual text files, but Korp gave me TSV file. It's not that I'm complaining since it also had lemmatized forms of original forum messages, but this requires additional process to create an individual text file for each forum message in the TSV file. The TSV file's messages can be easily turned into individual files with following code:
import pandas as pd
df = pd.read_csv("datasetti.tsv")
sanelut = list(df["text"])
for i in range(len(sanelut)):
f = codecs.open('"C:/Users/KoodiKoira/nertagger/finnish-tagtools-1.3.2/text_input/tc' + str(i+1) + '.txt', "w", "utf-8")
f.write(korjatut[i])
f.close()
Running FiNER¶
Now the files are ready for FiNER (short for Finnish Named Entity Tagger) and its additional package finer-utilities, which can be used to linearize tagged text documents. As previously mentioned, FiNER requires Linux environment, so I had to dual boot my laptop with a Linux distro. Dual booting was an easy process, but when it comes to ease of use, don't believe the Linux enthusiasts. Even Ubuntu isn't nearly as user friendly it is claimed, and that's supposed to be the most streamlined distro! It can be quite frustrating to manually install drivers for each individual software and hardware you use, alongside bugs in basic software programs, such as notepad. But maybe these things are only a problem for a novice coder such as myself, so I better quit my whining and switch back on the topic at hand.
I recommend downloading the FiNER 1.3.2 version, since I've experienced problems with the newer 1.4.0. Once the folder is downloaded, the user has to change the working directory to the downloaded finnish-tagtools folder and run sudo make install command. I've also added two folders into the finnish-tagtools folder, one for the input text files and one for output text files. Once the environment was established, I created a .sh script file that processes all text files in the input files folder. I included following bash commands, that in essence tags all the text files in the input folder and linearizes the tagged text file into easily readable format:
for file in ./input_texts/*.txt
do
finnish-nertag --show-nested < ./input_texts/\"${file##*/}\" > ./output_texts/$(basename -- \"tagged_${file##*/}\")
sudo bash linearize.sh < ./output_texts/\"tagged_${file##*/}\" > ./output_texts/$(basename -- \"tag_lin_${file##*/}\")
echo \"$(basename -- $file) processed\" # prints the last processed file in the console
done;
--show nested parameter adds multiple tags for one named entity if it has multiple meanings. One can also add --show-analyses parameter into the script to receive additional analyses, and morphological tags, but my dataset was already lemmatized that wasn't necessary, since I was only interested in named entity tags. I took quite a while to process all 3000 text documents, but it was done without major errors and now documents are ready for further analysis and processing!
Analyzing and Post-processing Tagged Text Documents¶
I wrote some code that creates simple UI for navigating and counting the frequency of each named entity tag from processed text documents. The code is available here, so I'll just show what it looks like.
With BeautifulSoup 4 Python package, it will be easy to pick each tag for processing, either for pseudonymizing or highlighting. I used following scripts to create both pseudonymized and highlighted text documents.
Tag Highlighting¶
from bs4 import BeautifulSoup
import os
tags = ["EnamexLocPpl", "EnamexLocGpl", "EnamexLocStr", "EnamexLocFnc", "EnamexLocAst",
"EnamexOrgPlt", "EnamexOrgClt", "EnamexOrgTvr", "EnamexOrgFin", "EnamexOrgEdu",
"EnamexOrgAth", "EnamexOrgCrp", "EnamexPrsHum", "EnamexPrsAnm", "EnamexPrsMyt",
"EnamexPrsTit", "EnamexProXxx", "EnamexEvtXxx", "TimexTmeDat", "TimexTmeHrm",
"NumexMsrXxx", "NumexMsrCur"]
PATH = "text_outputs/"
files = []
for file in os.listdir(PATH):
if os.path.isfile(os.path.join(PATH,file)) and 'tag_lin_tc' in file:
files.append(file)
textlist = []
for file in files:
soup = BeautifulSoup(open(PATH + file, 'r', encoding='utf-8'))
textlist += soup
f = open('KorpHighlighted.html', 'w+', encoding="utf-8")
f.write("<html>\n<head>\n<style>\n\ntable, th, td {\n border: 1px solid black;\n}\nenamexprshum {\n color:red;font-weight: bold;}\nenamexlocppl, enamexlocgpl {\n color:blue;font-weight: bold;}\nenamexlocstr, enamexlocfnc, enamexlocast, enamexorgplt, enamexorgclt, enamexorgtvr, enamexorgfin, enamexorgedu, enamexorgath, enamexorgcrp, enamexprsanm, enamexprsanm, enamexprsmyt, enamexprstit, enamexproxxx, enamexevtxxx, timextmedat, timextmehrm, numexmsrxxx, numexmsrcur{\n color:green;font-weight: bold;}\n</style>")
f.write('<table style="width:100%">')
f.write('<tr><th>id</th><th>Text</th></tr>')
for i in range(len(textlist)):
f.write('<tr><td>sample_text_' + str(i+1) + '</td><td>' + str(textlist[i]) + '</td></tr>')
f.write('</table>\n</body>\n</html>')
f.close()
Small sample of the Suomi24 corpus with plain FiNER tagged elements highlighted in colors:
| id | Tagged and lemmatized text |
|---|---|
| sample_text_229 |
erityinen mieli tulla , että |
| sample_text_230 | tämä vuoksi me olla jälleen kerta painottaa , kuten |
| sample_text_231 | me olla kuitenkin varmistaa , että tämä |
Tag Pseudonymization¶
textlist2 = []
for file in files:
soup = BeautifulSoup(open(PATH + file, 'r', encoding='utf-8'))
for etunimi in soup.findAll(tags[12].lower()): # tags[12] = EnamexPrsHum
etunimi.string = 'NAMECODE_' + "[" + str(file) + "]" # This is document specific pseudonymization, not individual name specific!
for paikannimi in soup.findAll(tags[0].lower()): # tags[12] = EnamexLocPpl
paikannimi.string = 'LOCATIONCODE_' + "[" + str(file) + "]"
for aluenimi in soup.findAll(tags[1].lower()): # tags[12] = EnamexLocGpl
aluenimi.string = 'AREACODE_' + "[" + str(file) + "]"
textlist2 += soup
f = open('KorpPseydonymized.html', 'w+', encoding="utf-8")
f.write("<html>\n<head>\n<style>\n\ntable, th, td {\n border: 1px solid black;\n}\nenamexprshum {\n color:red;font-weight: bold;}\nenamexlocppl, enamexlocgpl {\n color:blue;font-weight: bold;}</style>")
f.write('<table style="width:100%">')
f.write('<tr><th>id</th><th>Text</th></tr>')
for i in range(len(textlist2)):
f.write('<tr><td>sample_text_' + str(i+1) + '</td><td>' + str(textlist2[i]) + '</td></tr>')
f.write('</table>\n</body>\n</html>')
f.close()
Small sample of the Suomi24 corpus with pseudonymized FiNER tagged elements highlighted in colors:
| id | Tagged and lemmatized text |
|---|---|
| sample_text_229 | erityinen mieli tulla , että |
| sample_text_230 | tämä vuoksi me olla jälleen kerta painottaa , kuten |
| sample_text_231 | me olla kuitenkin varmistaa , että tämä |