Some exposition
Since graduating from my masters studies and condemning myself to toil on a payroll, I haven't had much inspiration to write any new blogs about data-analytic applications. Not that I haven't had any personal projects here and there, but nothing that I would've considered interesting enough to write about. On the other hand, if I had anything more aspirational in mind, it would inevitably fall apart and come to nothing due to lack of knowledge, time, energy and resources. Despite my technosceptisism, I started testing the capabilities of commercial large language models (LLMs), mainly Microsoft Copilot for project theorizing, conseptualizing and preliminary building and Google Gemini for fixing, patching and filing the rough edges.
My LLM use began when my workplace bought a Copilot license, which I could put to use as an AI assistant for work. However my main expertize depends of deep knowledge of regional healthcare service systems and healthcare organizational networks and Copilot lacks the context for this niche and interpersonal domain. This narrows Copilot's range of usefulness to summarizing public documents and helping to build more complex SQL queries in niche research dataset collection tasks. Due to the lack of direct use cases in addition to my initial and still somewhat persistent sceptical attitude towards LLMs, I ignored them for a good year, until lately I came across very interesting idea of running LLMs locally with Ollama and fine-tuning LLMs for specific tasks.
Ollama has been around a good while, so I'm a latecomer. I found it very intriguing that I'd be able to download any huggingface model and run it on my laptop! As I use a debian distro as my operating system, the installation process reguired only a couple console commands: an install command (curl -fsSL https://ollama.com/install.sh | sh), a pull command from Ollama’s LLM-library to download a LLM (ollama pull [model_name]) and a run command (ollama run [model_name]) to start chatting with the downloaded LLM. So essentially the actual process of downloading, installing and running LLMs took only 5-10 minutes (depending on your internet speed and the size of LLM you downloaded).
$ ollama run deepseek-r1:1.5b
Write a prompt: My knee hurts
It sounds like you might be experiencing pain related to your knee. If
this is happening on its own or if the pain gets worse, it could be
something serious or a sign of an injury. Here are some steps you can
take:
Rest: Try to lie down and get plenty of rest. Avoid sitting for
long periods.
Stretching: Engage in stretching exercises that target the knee
area. This might include stretches for the quadriceps (to strengthen your
core) or leg muscles (to improve flexibility)...
$ ollama pull deepseek-r1:1.5b
$ ollama ls
NAME ID SIZE MODIFIED
deepseek-r1:1.5b e0979632db5a 1.1 GB 10 seconds ago
medllama2:latest a53737ec0c72 3.8 GB 2 months ago
meditron:latest ad11a6250f54 3.8 GB 2 months ago
html-model:latest bc280c500d09 2.3 GB 3 months ago
deepseek-r1:8b 6995872bfe4c 5.2 GB 3 months ago
phi4:latest ac896e5b8b34 9.1 GB 3 months ago
... and so on and so on ...
Anyhow, my little toaster was powerful enough to chat with only the smallest LLMs, more aptly named: small language models (SLMs), which didn’t have enough context to provide any meaningful insight in a thoughtful conversation. However, a SLM could be implemented in more specialized tasks with greater computational efficiency compared to LLMs. What if I wished to extract, enrich or classify unstructured text data using these SLMs? They could perhaps give some kind of an answer depending on the model and prompt but they would need some example inputs and outputs to so that they can provide results with sufficient consistency. Experimenting with this would be interesting and perhaps even useful indeed, but it will require aforementioned unstructured text data for fine-tuning and testing!
Creating a randomly generated patient cohort
So, I have a new project in mind, which is to create a SLM that can detect and extract clinically relevant information from unstructured free-text electronic health recods (EHRs) and store it in structured format, in this case JSON. I could have used Copilot to scrape the web and provide me with necessary material, but I wanted to create the dataset myself! So I have this in mind: download a bunch of code systems related to personal names, place names, diagnoses, medications and measurements. Then use these clinical code and name datasets, along with random number generators with added weights to create a structured dataset of patient visit.
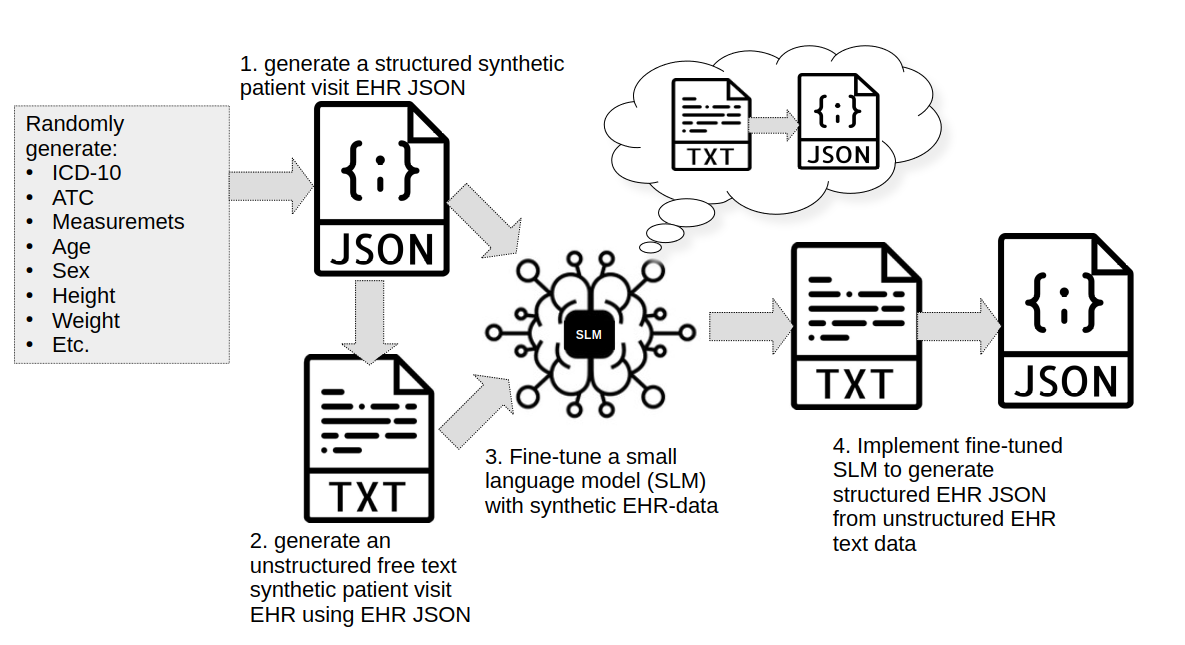 Image 1. The plan on (1.) creating a synthetic EHR JSON dataset using python randomization package along with diagnosis (ICD-10), medicine (ATC) and other clinical features using lookup lists formed from open data sources. After EHR JSON dataset is created (2.) it will be used to generate an unstructured free text synthetic patient visit EHR using open data sources and python randomization package. (3.) These two datasets will be used to fine-tune (free-text EHR as input and EHR json as output) a small language model (SLM) to detect and mine clinically relevant information from free-text EHRs into structured JSON-format. (4.) Fine-tuned SLM will be tested and implemented in generating structured EHRs from unstructured free-text EHRs. Only parts 1. and 2. will be demonstrated in this blog.
Image 1. The plan on (1.) creating a synthetic EHR JSON dataset using python randomization package along with diagnosis (ICD-10), medicine (ATC) and other clinical features using lookup lists formed from open data sources. After EHR JSON dataset is created (2.) it will be used to generate an unstructured free text synthetic patient visit EHR using open data sources and python randomization package. (3.) These two datasets will be used to fine-tune (free-text EHR as input and EHR json as output) a small language model (SLM) to detect and mine clinically relevant information from free-text EHRs into structured JSON-format. (4.) Fine-tuned SLM will be tested and implemented in generating structured EHRs from unstructured free-text EHRs. Only parts 1. and 2. will be demonstrated in this blog.
So the patient visit information will be created and stored in Patient and PatientRegistry classes as follows:
class Patient:
"""
A class to represent a patient.
"""
def __init__(self, id, visit_datetime, firstname, lastname, age, gender, height, weight,
bmi, diagnoses_old, diagnoses_new, medications, measurements, attitude, cognition, county, smokes):
"""
Initialize a Patient instance.
"""
self.id = id
self.visit_datetime = visit_datetime
self.firstname = firstname
self.lastname = lastname
self.age = age
self.gender = gender
self.height = height
self.weight = weight
self.bmi = bmi
self.diagnoses_old = diagnoses_old
self.diagnoses_new = diagnoses_new
self.medications = medications
self.measurements = measurements
self.attitude = attitude
self.cognition = cognition
self.homecounty = county
self.smokes = smokes
def __str__(self):
"""
String representation of the Patient object.
"""
return f"Patient(id: {self.id}, Visit Datetime: {self.visit_datetime}, Name: {self.firstname}, Last name: {self.lastname}, Age: {self.age}, Gender: {self.gender}, Height: {self.height}, Weight: {self.weight}, BMI: {self.bmi}, ICD10 in this session: {self.diagnoses_new}, ICD10-history: {self.diagnoses_old}, Medications: {self.medications}, Measurements: {self.measurements}, Attitude: {self.attitude}, Cognition: {self.cognition}, Homecounty: {self.homecounty}, Smoking status: {self.smokes})"
class PatientRegistry:
"""
A class to manage a collection of patients.
"""
def __init__(self):
"""
Initialize an empty patient registry.
"""
self.patients = []
def add_patient(self, patient):
"""
Add a patient to the registry.
:param patient: A Patient object
"""
if not isinstance(patient, Patient):
raise TypeError("Only Patient objects can be added.")
self.patients.append(patient)
def patient_generator(self):
"""
A generator to iterate over patients in the registry.
"""
for patient in self.patients:
yield patient
I also had some procedure codes but forgot to add them into the patient generation code. Despite that current number of variables should be sufficient enough to demonstrate this process. Also, you can see how the peculiar way the code is commented. It being clearly not my style betrays my use of LLMs, which I implemented to help me build a foundation for my project. To compare this to my own handiwork, see the code cell below how I've written the function calls that actually create the patients in registry:
num_patients = 2000 # here we create 2000 patients
# Create a patient registry
registry = PatientRegistry()
for i in range(num_patients):
# random id code to identify patient
id = ''.join(random.SystemRandom().choice(string.ascii_uppercase + string.digits) for _ in range(10))
sex = random.choice(["Male", "Female"])
if sex == "Male":
firstname = random.choice(menimet.tolist())
else:
firstname = random.choice(nenimet.tolist())
age = random.randint(18,80) # age is determined by a random integer between 18 and 80
# I could show you how the diagnoses are determined, but I'll spare you from seeing my spaghetti code
# let's just say that it is an amalgam of random samplings and if-else clauses /
# and desperately held together by try-except statements
old_diagnoses=generate_diagnosis_history(age, sex, 0)
new_diagnoses=generate_new_diagnosis(old_diagnoses['ICD10'], age, sex)
# medications is a funny one, if I recall correctly the str check is for statement that /
# there are no registered medications for the patients, but I removed that string statement /
# but I forgot to take that check in mind in this patient medication generation function /
# but hey! It works nevertheless!
if type(old_diagnoses["ICD10"]) == str and type(new_diagnoses["ICD10"]) == str:
medications = generate_medication([])
elif type(old_diagnoses["ICD10"]) == list and type(new_diagnoses["ICD10"]) == str:
medications = generate_medication(old_diagnoses["ICD10"])
elif type(old_diagnoses["ICD10"]) == str and type(new_diagnoses["ICD10"]) == list:
medications = generate_medication(new_diagnoses["ICD10"])
else:
medications = generate_medication(old_diagnoses["ICD10"]+new_diagnoses["ICD10"])
bmi = generate_height_weight(sex, h_hist, w_hist)
# Add some patients
registry.add_patient(Patient(id=id,
visit_datetime=random_date("1-1-2000 1:30:00", "1-1-2025 4:50:59", random.random()),
firstname=firstname, lastname=random.choice(snimet.tolist()),
age=age, gender=sex, height=bmi["height_cm"], weight=bmi["weight_kg"],
bmi=bmi["bmi"], diagnoses_old=old_diagnoses, diagnoses_new=new_diagnoses,
medications=medications, measurements=generate_measurements(),
attitude=random.choice(mental_states), cognition=random.choice(cognitions),
county=random.choice(counties), smokes=random.choice(smoking)
)
)
# then we'll save the synthetic patient registry in json format
import json
with open('synthetic_patients.json', 'w') as outfile:
outfile.write(json.dumps({"patients":[patient.__dict__ for patient in registry.patient_generator()]}, indent=4, ensure_ascii=False))
outfile.close()
Characterizing the dataset
Now that I've created the synthetic patients dataset. Let's check out the contents.
import json
# read in created dataset and pick a random patient to see what kind of informat\u00f6ion we have created
with open('synthetic_patients.json') as f:
patients = json.load(f)
print(json.dumps(patients['patients'][2], indent=4))
{
"id": "VLWFRNRQ9W",
"visit_datetime": "27-03-2008 12:55:23",
"firstname": "Lumi-Tuuli",
"lastname": "Korhola",
"age": 26,
"gender": "Female",
"height": 167,
"weight": 61,
"bmi": 21.87,
"diagnoses_old": {
"ICD10": [
"K50014"
],
"ICD10 description": [
"K50014 - Crohn's disease of large intestine with rectal bleeding"
]
},
"diagnoses_new": {
"ICD10": [
"K638212",
"K50014"
],
"ICD10 description": [
"K638212 - Perforation of intestine (nontraumatic)",
"K50014 - Crohn's disease of small intestine with abscess"
]
},
"medications": [
{
"ATC": "A03FA01",
"Indication": "K50014",
"Drug_name": "metoclopramide",
"dose": 30.0,
"unit": "mg",
"form": "P",
"note": NaN
},
{
"ATC": "A04AA01",
"Indication": "K50014",
"Drug_name": "ondansetron",
"dose": 16.0,
"unit": "mg",
"form": "R",
"note": NaN
},
{
"ATC": "A07EC01",
"Indication": "K50014",
"Drug_name": "sulfasalazine",
"dose": 2.0,
"unit": "g",
"form": "R",
"note": NaN
}
],
"measurements": [
{
"Measurement": "S -RaiheiE",
"Value": 0.24,
"Unit": "kU/l"
},
{
"Measurement": "S -21-OHAb",
"Value": 6.5,
"Unit": "Indeksi"
},
{
"Measurement": "P -Glkg",
"Value": 191.62,
"Unit": "ng/l"
},
{
"Measurement": "S -AGM1AbG",
"Value": 4.96,
"Unit": "Indeksi"
}
],
"attitude": "anxious",
"cognition": "stupor",
"homecounty": "Kitee",
"smokes": "Never smoked"
}
# let's also get a nice list of keys in the JSON dictionary
patients['patients'][2].keys()
dict_keys(['id', 'visit_datetime', 'firstname', 'lastname', 'age', 'gender', 'height', 'weight', 'bmi', 'diagnoses_old', 'diagnoses_new', 'medications', 'measurements', 'attitude', 'cognition', 'homecounty', 'smokes'])
Nice, so we have quite a lot of data here, each variable containing some data pertaining to the patient, such as name, residence, physiological, diagnostic, medicinal, psychological and lifestyle information. Let's try to visualize the dataset! Since the json schema contains nested lists and dictionaries, I'll have to save basic unnested items into a "wide" dataframe, that features a column for each item value and a "long" dataframe that has a "variable" column and "value" column that presents each item as a row entry without its own dedicated column.
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
# first I'll create a wide dataset, that will store unnested items
keys = ['id', 'visit_datetime', 'firstname', 'lastname',
'age', 'gender', 'height', 'weight', 'bmi',
'attitude', 'cognition', 'homecounty', 'smokes']
patient_wide = pd.DataFrame(columns=keys)
for patient in patients['patients']:
patient_wide = pd.concat([patient_wide, pd.DataFrame([{key: patient[key] for key in keys}])], ignore_index=True)
# then I'll create a long dataset, that will store nested items
keys2 = ['diagnoses_new', 'diagnoses_old', 'measurements', 'medications']
patient_long = pd.DataFrame(columns=['visit_datetime', 'id', 'variable', 'code', 'value', 'unit'])
# I must detect is the item a list or a dictionary and then iterate and store each item in that list/dict
for patient in patients['patients']:
dftemp = pd.DataFrame(columns=patient_wide.columns)
for key in keys2:
if key == 'diagnoses_new' or key == 'diagnoses_old':
if len(patient[key]['ICD10']) != 0:
for i in range(len(patient[key]['ICD10'])):
patient_long = pd.concat([patient_long, pd.DataFrame([{
'visit_datetime': patient['visit_datetime'],
'id': patient['id'],
'variable': key,
'code': patient[key]['ICD10'][i],
'value': None, 'unit': None }])], ignore_index=True)
else:
if len(patient[key])!=0:
if key == 'measurements':
for i in range(len(patient[key])):
try:
patient_long = pd.concat([patient_long, pd.DataFrame([{
'visit_datetime': patient['visit_datetime'], 'id': patient['id'],
'variable': key, 'code': patient[key][i]['Measurement'],
'value': patient[key][i]['Value'],
'unit': patient[key][i]['Unit']}])], ignore_index=True)
except:
# there ended up a few empty dictionaries that break the loop, so I added this as a remedy
pass
else:
for i in range(len(patient[key])):
patient_long = pd.concat([patient_long, pd.DataFrame([{
'visit_datetime': patient['visit_datetime'], 'id': patient['id'],
'variable': key, 'code': patient[key][i]['ATC'],
'value': patient[key][i]['dose'],
'unit': patient[key][i]['unit']}])], ignore_index=True)
All done! Let's see what the data looks like.
patient_wide.head()
| id | visit_datetime | firstname | lastname | age | gender | height | weight | bmi | attitude | cognition | homecounty | smokes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 61AT14FYFP | 04-07-2021 11:47:29 | Jawad | Löytönen | 69 | Male | 186 | 74 | 21.39 | unemotional | delirious | Kuopio | Never smoked |
| 1 | K8R9F90HZ2 | 15-08-2022 07:33:54 | Jiko | Jäppinen | 75 | Male | 176 | 72 | 23.24 | sad | delirious | Tohmajärvi | Has quit smoking |
| 2 | VLWFRNRQ9W | 27-03-2008 12:55:23 | Lumi-Tuuli | Korhola | 26 | Female | 167 | 61 | 21.87 | anxious | stupor | Kitee | Never smoked |
| 3 | OE58MF3147 | 14-01-2014 07:32:02 | Hannele | Kortesluoma | 56 | Female | 169 | 55 | 19.26 | unemotional | delirious | Juva | Smokes currently |
| 4 | OACDL8PNWD | 07-01-2010 02:47:59 | Pulmu | Kulju | 66 | Female | 163 | 55 | 20.70 | sad | unfocused | Kontiolahti | Never smoked |
patient_long.head()
| visit_datetime | id | variable | code | value | unit | |
|---|---|---|---|---|---|---|
| 0 | 04-07-2021 11:47:29 | 61AT14FYFP | diagnoses_new | J1000 | NaN | None |
| 1 | 04-07-2021 11:47:29 | 61AT14FYFP | diagnoses_new | M5481 | NaN | None |
| 2 | 04-07-2021 11:47:29 | 61AT14FYFP | diagnoses_new | H4050X4 | NaN | None |
| 3 | 04-07-2021 11:47:29 | 61AT14FYFP | diagnoses_old | H53453 | NaN | None |
| 4 | 04-07-2021 11:47:29 | 61AT14FYFP | diagnoses_old | A529 | NaN | None |
Seems sensible enough, let's characterize the data by visualizing it with matplotlib and seaborn.
from matplotlib import pyplot as plt
import seaborn as sns
#I'd like to see cognitive states by age, but it is a bit too messy to use every age as category, so I'll group them
bins = [17, 30, 50, 70, 90] # Intervals: 18-30, 31-50, 51-70, 71-90
labels = ['18-30', '31-50', '51-70', '71-90']
# 2. Create the Age Group column
patient_wide['AgeGroup'] = pd.cut(patient_wide['age'], bins=bins, labels=labels, right=True)
sns.set_theme()
p=sns.catplot(
patient_wide, kind="bar",
x="AgeGroup", y="bmi", hue='gender', errorbar=None,
height=3, aspect=2, estimator='mean'
)
p.fig.suptitle('BMI by age and gender')
Text(0.5, 0.98, 'BMI by age and gender')
There doesn't seem to be any variance in bmi either by age, nor gender. Let's check the number of psychiatric items.
p=sns.countplot(patient_wide, x='attitude', hue='gender')
p.set_title('Emotional states by gender')
Text(0.5, 1.0, 'Emotional states by gender')
sns.set_theme(rc={'figure.figsize':(7,5)})
p=sns.countplot(patient_wide, x='AgeGroup', hue='cognition')
p.set_title('Cognitive states by age')
Text(0.5, 1.0, 'Cognitive states by age')
sns.set_theme(rc={'figure.figsize':(6,4)})
p=sns.countplot(patient_wide, x='smokes', hue='gender')
p.set_title('Smoking status by gender')
Text(0.5, 1.0, 'Smoking status by gender')
sns.set_theme(rc={'figure.figsize':(7,5)})
p=sns.countplot(patient_wide, x='smokes', hue='AgeGroup')
p.set_title('Smoking status by age')
Text(0.5, 1.0, 'Smoking status by age')
import numpy as np
sns.set_theme(rc={'figure.figsize':(10,6)})
p=sns.countplot(patient_wide, x='height', hue='gender')
p.set_xticklabels(labels=np.sort(patient_wide['height'].unique()),rotation=90)
p.set_title('Height by gender')
Text(0.5, 1.0, 'Height by gender')
Let's see the number of old and new diagnoses, measurements and medications per patient.
lm = []
la = []
for variable in patient_long['variable'].unique():
print(f"{variable}: {float(patient_long[patient_long['variable']==variable].groupby('id').size().mean())}")
la.append(float(patient_long[patient_long['variable']==variable].groupby('id').size().mean()))
lm.append(variable)
plotdf = pd.DataFrame({'variable': lm, 'mean_value': la})
sns.set_theme(rc={'figure.figsize':(6,4)})
p=sns.barplot(plotdf, x='variable', y='mean_value', estimator='sum')
p.set_title('Average number of diagnoses, measurements and medications per patient')
diagnoses_new: 2.363 diagnoses_old: 1.8701456310679612 measurements: 2.949275362318841 medications: 3.636175710594315
Text(0.5, 1.0, 'Average number of diagnoses, measurements and medications per patient')
# Creating a high-level ICD10 parent code for plot readability
patient_long['parent_code'] = patient_long.code.apply(lambda x: x[:2])
# Also adding age and gender information for grouping
patient_long = patient_long.merge(patient_wide[['id','AgeGroup', 'gender']], on = 'id', how='left')
sns.set_theme(rc={'figure.figsize':(10,5)})
p=sns.countplot(patient_long[((patient_long['variable'] == 'diagnoses_old')|(patient_long['variable'] == 'diagnoses_new')) & (patient_long['parent_code'].map(patient_long['parent_code'].value_counts()) > 100)]
, x='parent_code', hue='gender')
#p.set_xticklabels(labels=np.sort(patient_long['parent_code'].unique()),rotation=90)
p.set_title('Number of diagnoses by ICD10 parent code and by gender')
Text(0.5, 1.0, 'Number of diagnoses by ICD10 parent code and by gender')
sns.set_theme(rc={'figure.figsize':(15,5)})
p=sns.countplot(patient_long[((patient_long['variable'] == 'diagnoses_old')|(patient_long['variable'] == 'diagnoses_new')) & (patient_long['parent_code'].map(patient_long['parent_code'].value_counts()) > 100)]
, x='parent_code', hue='AgeGroup')
#p.set_xticklabels(labels=np.sort(patient_long['parent_code'].unique()),rotation=90)
p.set_title('Number of diagnoses by ICD10 parent code and by age group')
Text(0.5, 1.0, 'Number of diagnoses by ICD10 parent code and by age group')
I added some weights in diagnosis randomizer based on age and gender. I hoped that those weights had a little more impact, but there still is some observable variance.
Is there really anything else to say? It is just a boring, mostly balanced dataset without any significant variance in data, because I didn't intend this dataset to have any differing trends, nor outliers either. This dataset is intended for few specific, non-statistical information extraction and classification cases.
Creating synthetic EHR "unstructured" text samples
Now that we have our patient cohort, we can use it to generate a "unstructured" text dataset to fine-tune a language model. I mention unstructured in quotations because I will use sentence samples of free text that I will shuffle and stitch together as a patchwork, which I then will corrupt a little bit. This synthetic text data will still contain certain degree of structurality which won't adequately represent true messiness of human-generated unstructured free text.
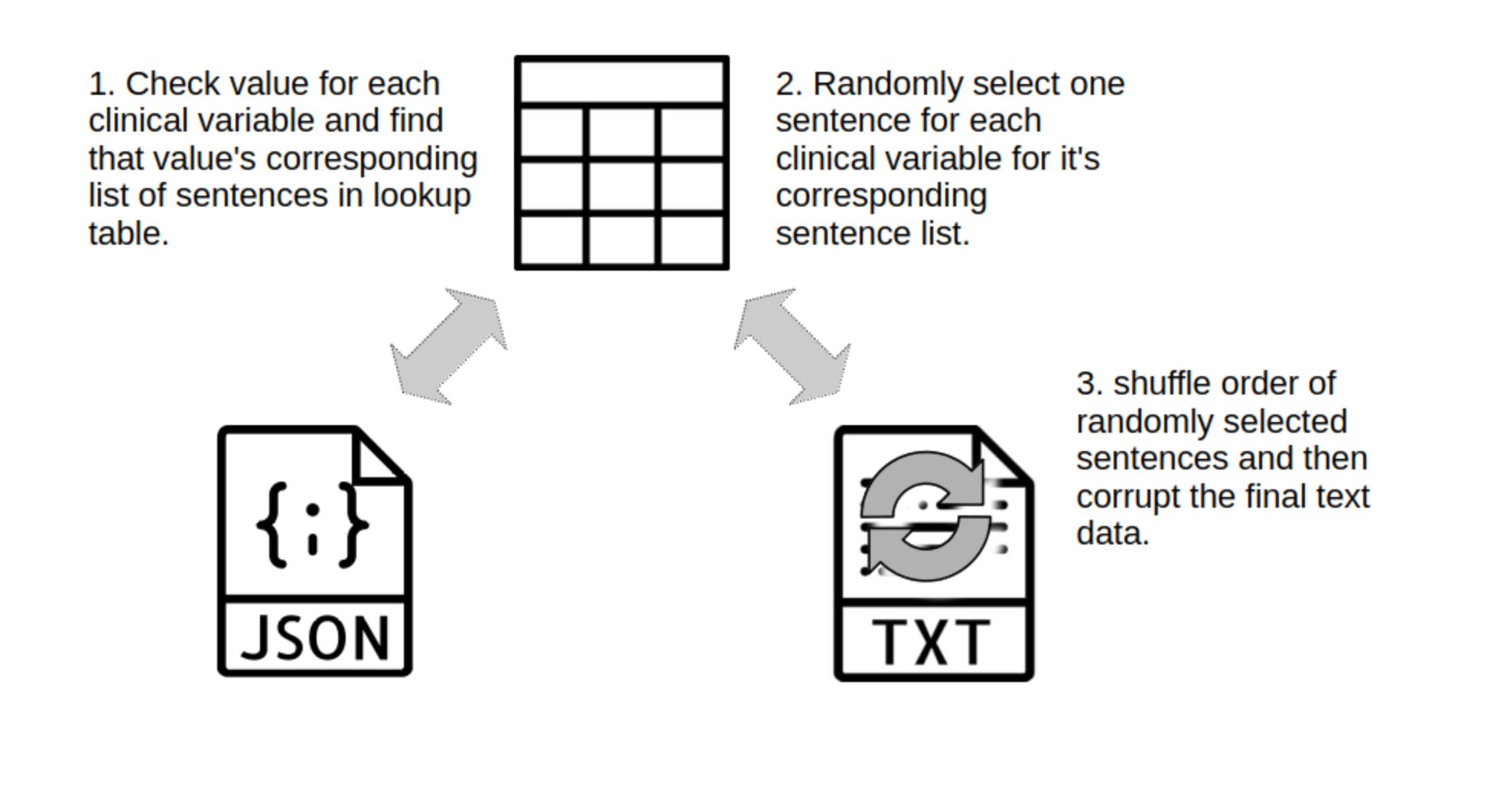 Image 2. Process of unstructured free-text EHR generation.
Image 2. Process of unstructured free-text EHR generation.from datasets import load_dataset
import corrupted_text # for text corruption
import pandas as pd
import numpy as np
import random
import time
from time import mktime
import json
import re
train_data = load_dataset("ag_news", split="train")["text"]
test_data = load_dataset("ag_news", split="test")["text"]
text_corruptor = corrupted_text.TextCorruptor(base_dataset=test_data + train_data, cache_dir=".mycache")
# load the synthetic patient dataset
with open('synthetic_patients.json') as f:
patients = json.load(f)
# load term and sentence datasets for text generation
beginning = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Introduction")
name = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Name declaration")
times = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Time declaration")
agesex = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Age Sex declaration")
hweight = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Height Weight declaration")
bmi = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="BMI declaration")
homecounty = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Homecounty declaration")
attitude = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Attitude declaration")
cognition = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Cognition declaration")
smoking = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Smoking status declaration")
ending = pd.read_excel("Aineistoa/Text_generator_choices.ods", sheet_name="Ending")
menimet = pd.read_excel("Aineistoa/etunimitilasto-2025-08-13-dvv.xlsx", sheet_name="Miehet ens", nrows=500)
nenimet = pd.read_excel("Aineistoa/etunimitilasto-2025-08-13-dvv.xlsx", sheet_name="Naiset ens", nrows=500)
nimet = pd.concat([menimet,nenimet]).reset_index()
snimet = pd.read_excel("Aineistoa/sukunimitilasto-2025-08-13-dvv.xlsx", sheet_name="Nimet", nrows=500)
The following step will be a little messy, but it adequately generates free text samples. Essentially it iterates through each patient and randomly selects a sentence sample from sentence lookup table from its corresponding clinical data item of the patient. Some of the sentences are embedded with references to patient data items, for which we'll need to interpret with eval(). After sentences are ready, they will be allocated into their sentence lists, representing different domain parts of clinical text. These sentence list items will be shuffled and then the lists will be joined together.
def generate_text(test):
i = random.randint(0,len(beginning)-1)
intr = eval(beginning["content"][i]) # always used as a start of the clinical text
t = random.choice(times["content"])
nd = random.choice(name["content"].tolist())
hw = random.choice(hweight["content"].tolist())
bm = random.choice(bmi[(bmi.lower < test["bmi"]) & (bmi.upper >= test["bmi"])]["content"].tolist())
asd = random.choice(agesex[agesex["value"]==test["gender"]]["content"].tolist())
hcd = random.choice(homecounty["content"].tolist())
ad = random.choice(attitude[attitude["value"]==test["attitude"]]["content"].tolist())
cd = random.choice(cognition[cognition["value"]==test["cognition"]]["content"].tolist())
sd = random.choice(smoking[smoking["value"]==test["smokes"]]["content"].tolist()).replace('“','"').replace('”','"')
mes = "Measurements: "
for measurement in test["measurements"]:
try:
mes += (measurement["Measurement"] + ": " + str(measurement["Value"]) + " " + measurement["Unit"] + ". ")
except:
pass
mes = mes[:-2] + ". "
meds = "Medications: "
for medication in test["medications"]:
meds += medication["Drug_name"]
meds += ", ATC: "
meds += medication["ATC"]
if pd.isna(medication["Indication"]) == False:
meds += ", indication: " + medication["Indication"]
if pd.isna(medication["dose"]) == False:
meds += ", dose: " + str(medication["dose"])
if pd.isna(medication["unit"]) == False:
meds += ", unit: " + str(medication["unit"])
if pd.isna(medication["form"]) == False:
meds += ", form: " + str(medication["form"])
if pd.isna(medication["note"]) == False:
meds += ", note: " + str(medication["note"])
meds += ", "
meds = meds[:-2] + ". "
if test['diagnoses_new']['ICD10'] != '':
icdd = "Current diagnoses: " + ''.join([x + ", " for x in test["diagnoses_new"]["ICD10 description"] ])[:-2] + ". "
else:
icdd = random.choice(["", "No new diagnoses. "])
if test['diagnoses_old']['ICD10'] != '':
icdhd = "Old diagnoses: " + ''.join([x + ", " for x in test["diagnoses_old"]["ICD10 description"] ])[:-2] + ". "
else:
icdhd = random.choice(["", "No previously recorded diagnoses. "])
end = random.choice(ending[ending["value"]==beginning["value"][i]]["content"].tolist()) # always used as an ending of the clinical text
a = eval(t) # time declaration
b = eval(nd) # name declaration
c = eval(hw) # height and weight declaration
d = eval(bm) # BMI declaration
e = eval(asd) # age and sex declaration
f = eval(hcd) # homecounty declaration
g = eval(ad) # attitude declaration
h = eval(cd) # cognition declaration
i = eval(sd) # smoking status declaration
j = mes
k = meds
l = icdd#.replace("None", "")
m = icdhd#.replace("None", "")
list1 = [a,b]
list2 = [c,d]
list3 = [e,f]
list4 = [g,h,i]
list5 = [j,k,l,m]
lista = ''.join([str(w) for w in random.sample(list1, len(list1))])+''.join([str(w) for w in random.sample(list2, len(list2))])+''.join([str(w) for w in random.sample(list3, len(list3))])+''.join([str(w) for w in random.sample(list4, len(list4))])+''.join([str(w) for w in random.sample(list5, len(list5))])
text = intr+lista+eval(end)
return text
# here we'll call the generate_text function for each patient and save generated texts in a list
idlista = [patients['patients'][i]['id'] for i in range(len(patients['patients']))]
textlist=[]
for patient in patients["patients"]:
textlist.append(generate_text(patient))
patient.pop('id', None) # here i'll just remove the id that'll I'll add to the dataset table as its own column
# after generating texts, I'll corrupt it to simulate typos and other writing errors into the text
corruption_severity = 0.05
corrupted_texts = text_corruptor.corrupt(textlist, severity=corruption_severity, seed=1)
import re
# The text corruption adds some artifacts which I don't want in the text so I'll clean it with regular expressions
def cleaner(text):
pattern1 = r'(?<=\d)\. (?=\d)'
pattern2 = r'(?<=\d) : (?=\d)'
pattern3 = r'(?<=.) : (?=.)'
pattern4 = r'(?<=.) - (?=.)'
pattern5 = r'(?<=.) \/ (?=.)'
pattern6 = r'(?<=.) \( (?=.)'
pattern7 = r'(?<=.) \) (?=.)'
pattern8 = r"(?<=.) \' (?=.)"
r1 = re.sub(pattern1, '.', text)
r2 = re.sub(pattern2, ':', r1)
r3 = re.sub(pattern3, ': ', r2)
r4 = re.sub(pattern4, '-', r3)
r5 = re.sub(pattern5, '/', r4)
r6 = re.sub(pattern6, ' (', r5)
r7 = re.sub(pattern7, ') ', r6)
r8 = r7.replace('"','').replace('’', "'")
r9 = re.sub(pattern8, "'", r8)
return r9
clean_c_texts = [cleaner(text) for text in corrupted_texts]
And there we have it! Now we have a synthetic EHR text dataset! Let's compare few examples between plain and corrupted texts.
| Plain text | Corrupted text |
| The patient was received in the outpatient department and escorted to the examination room. Baseline vitals were obtained per general medicine protocol. Patient's name: Lumi-Tuuli Korhola. Date and time of service 27-03-2008 12:55:23. Healthy body size. She is 167cm tall and weighs 61kg. Person is a female 26 years of age. Resides in Kitee. Patient appears nervous. Responds coherently to questions only through strained effort. Patient doesn't smoke. Measurements: S -RaiheiE: 0.24 kU/l. S -21-OHAb: 6.5 Indeksi. P -Glkg: 191.62 ng/l. S -AGM1AbG: 4.96 Indeksi. Old diagnoses: K50014 - Crohn's disease of large intestine with rectal bleeding. Medications: metoclopramide, ATC: A03FA01, indication: K50014, dose: 30.0, unit: mg, form: P, ondansetron, ATC: A04AA01, indication: K50014, dose: 16.0, unit: mg, form: R, sulfasalazine, ATC: A07EC01, indication: K50014, dose: 2.0, unit: g, form: R. Current diagnoses: K638212 - Perforation of intestine (nontraumatic), K50014 - Crohn's disease of small intestine with abscess. The patient was guided to the next available examination room. Intake staff completed all required preliminary checks. | The patient was received in tha outpatient department and escorted to the examination room. Baseline vitals were obtained per general medicine protocol. Patient's name: Lumi-Tuuli Korhola. Date and time of service 27-03-2008 12:55:23. Healthy body size. She is 167cm tall and weighs 61kg. Person is a female 26 years of age. Resides in Kitee. Patient appears nervous. Responds coherently to questions only through strained effort. Patient doesn't smoke. Measurements: S-RaiheiE: 0.24 kU/l. S-21-OHAb: 6.5 Indeksi. P-Glkg: 191.62 ng/l. S-AGM1AbG: 4.96 Indeksi. Old diagnoses: K50014-Crohn's disease of large intestine with rectal bleeding. Medications: metoclopramide, ATC: A03FA01, indication: K50014, dose: 30.0, unit: mg, form: P, ondansetron, ATC: A04AA01, indication: K50014, dose: 16.0, unit: mg, form: R, sulfasalazine, ATC: A07EC01, indication: K50014, dose: 2.0, unit: g, form: R. Current diagnoses: K638212-Perforation of intestine (nontraumatic ), K50014-Crohn's disease of small intestine withdraw abscess. The patient was guided to theft next available examination room. Intake staff completed all required preliminary checks. |
| A patient arrives with a referral from Dr. Klaus Suvanto. Name: Pulmu, last name: Kulju. Date and time of this recording 07-01-2010 02:47:59. Measured at height 163 cm and weight 55 kg during reception procedures. Age: 66 years. Sex: female. Indicates residence in Kontiolahti, documented at reception. No smoking. Patient's thoughts drift, and responses are tangential to the topic. Has a negative, dejected attitude. Medications: sodium phenylbutyrate, ATC: A16AX03, indication: R739, dose: 20.0, unit: g, form: O, eliglustat, ATC: A16AX10, indication: R739, dose: 0.168, unit: g, form: O. Measurements. Current diagnoses: F15180 - Other stimulant abuse with intoxication, unspecified, R739 - Hyperglycemia, unspecified, M13872 - Other microscopic hematuria. The patient completed the intake process and was instructed to await further instructions. No immediate interventions were required. | A patient arrives jith a referral from Dr. Klaus Suvanto. Name: Pulmu, last name: Kulju. Date and time of this recording 07-01-2010 02:47:59. Measured at height 163 cm and weight 55 kg during reception procedures. Age: 66 nears. Sex: female. Indicates residence in Kontiolahti, documented at reception. No smoking. Patient's thoughts drift, and responses are tangential to the topic. Has a negative, dejected attitude. Medications: sodium phenylbutyrate, ATC: A16AX03, indication: R739, dose: 20.0, unit: g, form: O, eliglustat, ATC: A16AX10, indication: R739, dose: 0.168, unit: g, form: O. Measurements. Current diagnoses: F15180-Other stimulant abuse with intoxication, unspecified, R739-Hyperglycemia, unspecified, M13872-Other microscopic hematuria. The patient completed the intake process and was instruments to await further instructions. No immediate interventions were required. |
| The patient was received in the outpatient department and escorted to the examination room. Baseline vitals were obtained per general medicine protocol. Name? Pirkko, and last name? Ruotsi. Her arrival at reception was at 25-02-2023 03:12:40. Somewhat malnourished. Her measured height is 179cm and a measured weight is 55kg. Home county noted as Kaavi during intake procedures. Sex: Female. Age: 65 years. Demonstrates difficulty organizing thoughts into coherent responses. Forgot to ask about patient's smoking habits.Presents with a subdued mood and speaks in a low, soft tone. Current diagnoses: F0789 - Other personality and behavioral disorders due to known physiological condition, S06329S - Contusion and laceration of left cerebrum with loss of consciousness of unspecified duration, sequela. Medications: isoflurane, ATC: N01AB06, indication: S06329S, ropivacaine, ATC: N01BB09, indication: S06329S. Measurements: S -rAspf2E: 0.23 kU/l. Li-Pyruv: 70.07 umol/l. S -BataatE: 0.26 kU/l. S -BopeAbM: 0.59 indeksi. S -RotseeE: 0.37 kU/l. Old diagnoses: S06329S - Injury of right internal carotid artery, intracranial portion, not elsewhere classified with loss of consciousness of 30 minutes or less, initial encounter, G8103 - Idiopathic sleep related nonobstructive alveolar hypoventilation, R532 - Hallucinations, unspecified, D500 - Sideropenic dysphagia, J203 - Acute bronchitis due to Hemophilus influenzae. The patient was guided to the next available examination room. Intake staff completed all required preliminary checks. | The patient was received in the outpatient department and escorted to the examination room. Baseline vitals wvre obtained per general medicine protocol. Name? Pirkko, and last name? Ruotsi. Her arrival at reception was at 25-02-2023 03:12:40. Somewhat malnourished. Her measure height is 179cm and a measured weight is 55kg. Home count noted as Kaavi during intake procedures. Sex: Female. Age: 65 years. Demonstrates difficulty organizing thoughts into coherent responses. Forgot to ask about patient's smoking habits. Presents with a subdued mood and speaks in a low, soft tone. Current diagnoses: F0789-Other personality and activity disorders due to known physiological condition, S06329S-Contusion and laceration of left cerebrum with loss of consciousness of unspecified duration, sequela. Medications: isoflurane, ATC: N01AB06, indication: S06329S, ropivacaine, ATC: N01BB09, indication: S06329S. Measurements: S-rAspf2E: 0.23 kU/l. Li-Pyruv: 70.07 umol/l. S-BataatE: 0.26 kU/l. S-BopeAbM: 0.59 indeksi. S-RotseeE: 0.37 kU/l. Old diagnoses: S06329S-Injury of right internal carolina artery, intracranial portion, not elsewhere classified with loss of consciousness of 30 minutes or less, initial encounter, G8103-Idiopathic sleep related nonobstructive alveolar hypoventilation, R532-Hallucinations, unspecified, D500-Sideropenic kysphagia, J203-Acute bronchitis due to Hemophilus influenzae. The patient was guided to the next available examination room. Intake staff completed all required preliminary check. |
As the reader can see, there is an observable general structure and pattern between these few text samples. If SLM would be deployed to production with this data, it most likely wouldn't perform that well, since it most likely overfits to the narrow variability presented in this dataset. Nevertheless, it could still prove useful for testing SLM fine-tuning.
# after that I'll save the dataset as an Excel-file to be used in fine-tuning
dftexts = pd.DataFrame({"id": idlista, "json": patients["patients"], "text original": textlist, "text corrupter": clean_c_texts})
dftexts.to_excel("synthetic_patient_texts.xlsx", index=None)
I was thinking of calling it here, but for the fun of it I'll throw in a freebie with a Doc2Vec clustering following the methods in this article. Here I'll represent each free-text EHR as a multidimensional vector, which I will then plot in two dimensional scatterplot using principal component analysis.
import pandas as pd
import numpy as np
import gensim
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
from gensim.models import Doc2Vec
train= pd.read_excel('synthetic_patient_texts.xlsx')
LabeledSentence1 = gensim.models.doc2vec.TaggedDocument
all_content_train = []
j=0
for em in train['text corrupter'].values:
all_content_train.append(LabeledSentence1(em,[j]))
j+=1
print("Number of texts processed: ", j)
Number of texts processed: 2000
# I could play around with parameters, but I'll just use the same parameters as within the article
d2v_model = Doc2Vec(all_content_train, vector_size = 100, window = 10, min_count = 500, workers=7, dm = 1,alpha=0.025, min_alpha=0.001)
d2v_model.train(all_content_train, total_examples=d2v_model.corpus_count, epochs=10, start_alpha=0.002, end_alpha=-0.016)
# I'll throw in 3 kmeans clusters and see what'll I get
kmeans_model = KMeans(n_clusters=3, init='k-means++', max_iter=100)
X = kmeans_model.fit(d2v_model.dv.vectors)
labels=kmeans_model.labels_.tolist()
l = kmeans_model.fit_predict(d2v_model.dv.vectors)
pca = PCA(n_components=2).fit(d2v_model.dv.vectors)
datapoint = pca.transform(d2v_model.dv.vectors)
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=(7,5))
label1 = ["#008000", "#0000FF", "#800080"]
color = [label1[i] for i in labels]
plt.scatter(datapoint[:, 0], datapoint[:, 1], c=color)
centroids = kmeans_model.cluster_centers_
centroidpoint = pca.transform(centroids)
plt.scatter(centroidpoint[:, 0], centroidpoint[:, 1], marker='^', s=150, c='#000000')
plt.show()
Don't let the pretty colors deceive you, it is just a one big blob. I haven't added enough consistently differing groups within the dataset so it all merges into one group. That doesn't matter though since this dataset's purpose isn't to be used to train a text classifier, but a text miner. But in future I'll use this method again to characterize another kind of dataset meant for text classification!
Discussion
In this blog post I have created a synthetic EHR dataset simulating 2000 patients, each with details from a single health care visit represented in JSON format and then created a synthetic EHR free-text dataset with same amount of samples. I will use this text data to fine-tune a SLM to extract clinical information from free-text EHRs back into structured JSON format from which I initially created the clinical text samples. Once that is done, I'll test the trained SLM with a subset of my dataset and compare its output with original json samples.
Data sources
- National codeserver for diagnoses, measurements and medications
- Digital and population data service agency for person and location names
- Using Electronic Patient Records to Discover Disease Correlations and Stratify Patient Cohorts for generating ICD-10 diagnosis comorbidity pairs
- And some other dataset sources which I have now forgotten 😐

