In my previous blog post I created a 2000 sample patient health care unit visitation EHR dataset represented both in JSON format. From this dataset I generated similar amount of clinical free-text samples which I will use along with JSON data to fine-tune a small language model (SLM) to extract clinical data from unstructured to structured format.
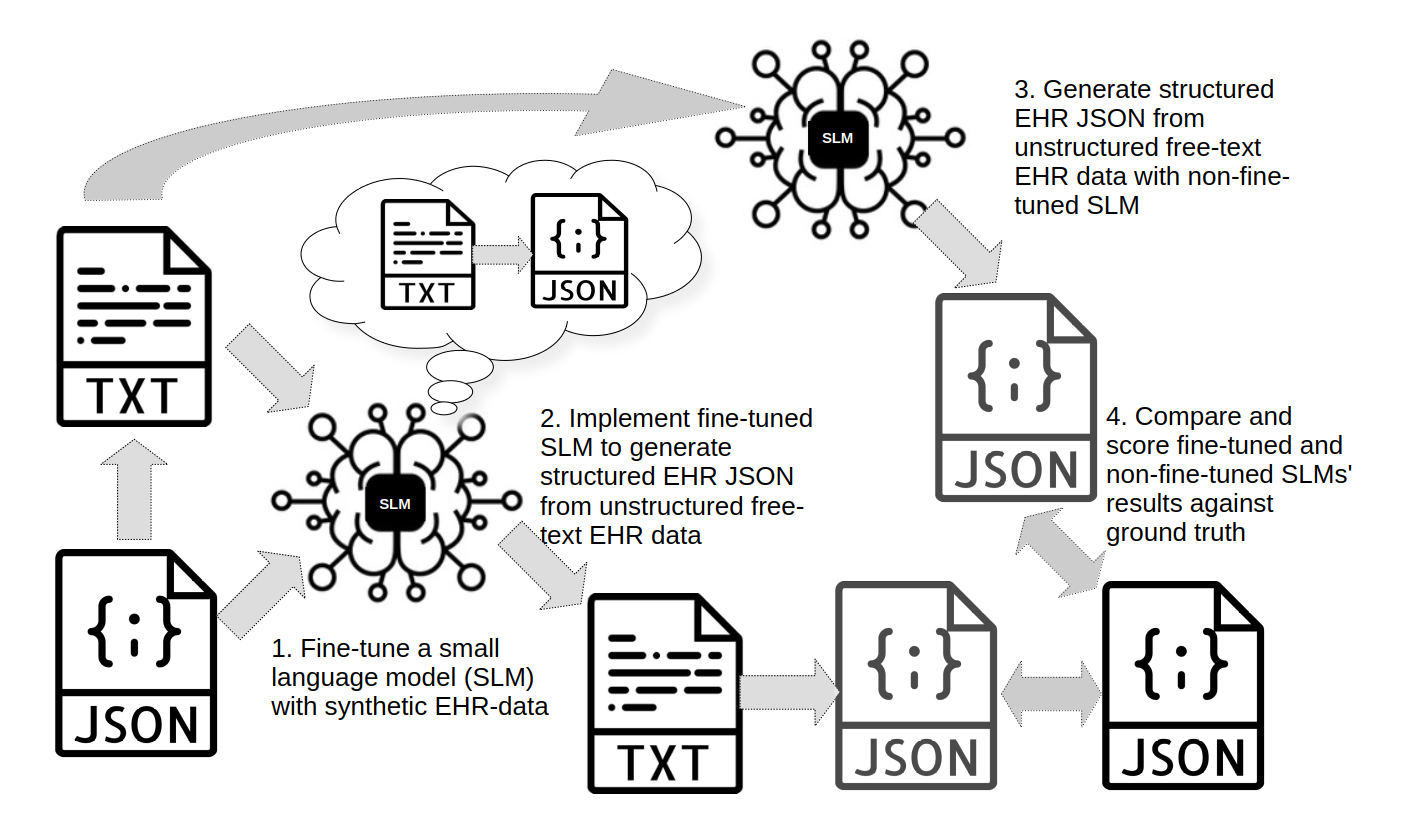
Figure 1. Abstraction of fine-tuning and test process implemented in this blog post. (1.) Previously prepared synthetic patient dataset and a text dataset derived from synthetic patient dataset will be used as training data to fine-tune Qwen3-4b-Instruct-2507 small learning model (SLM) to more efficiently extract clinically relevant information from unstructured free-text EHR data. 1600 samples will be used for training. (2.) Fine-tuned model will be used to produce structured clinical EHR from 100 samples of synthetic unstructured free-text EHR data and alongside this (3.) non-fine-tuned model also will produce structured EHR data from same test dataset. (4.) Results from fine-tuned and non-fine-tuned models will be tested and compared against test dataset's ground truth and improvement of model performance by fine-tuning will be evaluated based on these results.
In this example I will use Google Colab workspace for fine-tuning since it has connection to GPU runtime. Training data will use unstructured free text sample as a training input and structured JSON as example output. I arbitrarily deemed 1600 text samples of sufficient size for fine-tuning purposes. Once that is done, I will upload trained model to Google Drive and from there download the model to my local computer. In this part Google Gemini came in handy since there are quite a lot of moving parts under the hood of each language model, and each don't work excactly the same as the other. So I got this pipeline working no thanks to my lackluster knowledge of language models, but by trial and error and additional assistance by Google Gemini, which fixed small errors here and there.
Below is an example of an input and output, which will be used to fine-tune Qwen3-4b-Instruct-2507 model to enhance its performance in clinical text mining.
| Input | Output |
| The patient checkered in at reception and appeared stable and cooperative. Initial symptom review was conducted during intake. Recording started at 15-08-2022 07:33:54. Person's name: Jiko Jäppineh. Measuring height at 176cm and weight at 72kg. 75 years old male. Recently moved to Tohmajärvi. Is not in complete control of his mental faculties. Patient seems to be depressed. Quit smoking. Measurements. Current diagnoses: R12-Heartburn, R12-Heartburn. Old diagnoses: R12-Heartburn. Medications: ondansetron, ATC: A04AA01, indication: R12, dose: 16.0, unit: Mg, form: P, dexlansoprazole, ATC: A02BC06, indication: R12, dose: 30.0, unit: mg, form: O. The patient remained stable throughout intake and was prepared for physician assessment. Documentation was completed and forwarded. |
{ "age": 75, "attitude": "sad", "bmi": 23.24, "cognition": "delirious", "diagnoses_new": { "ICD10": [ "R12", "R12" ], "ICD10 description": [ "R12 - Heartburn", "R12 - Heartburn" ] }, "diagnoses_old": { "ICD10": [ "R12" ], "ICD10 description": [ "R12 - Heartburn" ] }, "firstname": "Jiko", "gender": "Male", "height": 176, "homecounty": "Tohmajärvi", "lastname": "Jäppinen", "measurements": [], "medications": [ { "ATC": "A04AA01", "Drug_name": "ondansetron", "Indication": "R12", "dose": 16.0, "form": "P", "note": null, "unit": "mg" }, { "ATC": "A02BC06", "Drug_name": "dexlansoprazole", "Indication": "R12", "dose": 30.0, "form": "O", "note": null, "unit": "mg" } ], "smokes": "Has quit smoking", "visit_datetime": "15-08-2022 07:33:54", "weight": 72 } |
Ignore the patient generator's diagnosis duplication artifact... moving on!
Preparing the training dataset and training parameters
Fine-tuning part of this project is performed in Google Colab, where I first must first download unsloth which is an open source tool that makes fine-tuning multiple times faster.
!pip install unsloth trl peft accelerate bitsandbytes
Installing dataset that has been uploaded into Colab's work repository.
import pandas as pd
df = pd.read_excel('sample_data/synthetic_patient_texts.xlsx')
dftrain = df.loc[0:1599] # 1600 samples for training
dftest = df.loc[1600:1999] # rest are left for test dataset
#dfval = df.loc[1800:1999] # due to memory constraints in Colab's free GPU runtime, using validation data wasn't an option :(
Checking that GPU resource is available.
# For GPU check
import torch
print(f"CUDA available: {torch.cuda.is_available()}")
print(f"GPU: {torch.cuda.get_device_name(0) if torch.cuda.is_available() else 'None'}")
Now let's download the model. After trying out different models and gradually decreasing model parameter size for each new model, I finally settled on this Qwen3-4B-Instruct-2507 model. The benchmarks indicated that this model could provide strong inference capabilities with not too many parameters, indicating that this could provide sufficient performance without being computationally overdemanding.
import unsloth
from unsloth import FastLanguageModel
#from transformers import AutoTokenizer, AutoModel
import torch
# would have preferred to use meditron this will have to do
model_name = "Qwen/Qwen3-4B-Instruct-2507"
max_seq_length = 1024 # Choose sequence length
dtype = None # Auto detection
# Load model and tokenizer
model, tokenizer = FastLanguageModel.from_pretrained(
model_name=model_name,
max_seq_length=max_seq_length,
dtype=dtype,
load_in_4bit=True # Changed to True to save memory
)
Here I'll reformat my dataset for training purposes. Each data entry contains both input data 'text corrupter' and output 'json'.
from datasets import Dataset
dftrain['data'] = dftrain.apply(lambda x: f"### Input: {x['text corrupter']}\n### Output: {x['json']}<|endoftext|>", axis=1)
#dfval['data'] = dfval.apply(lambda x: f"### Input: {x['text corrupter']}\n### Output: {x['json']}<|endoftext|>", axis=1)
# format training dataset
formatted_train_data = [input for input in dftrain['data']]
train_dataset = Dataset.from_dict({"text": formatted_train_data})
# format validation dataset
#formatted_eval_data = [input for input in dfval['data']]
#eval_dataset = Dataset.from_dict({"text": formatted_eval_data})
Due to GPU cluster's memory limitations I went with lowest possible QLoRA training parameters.
# Add LoRA adapters
model = FastLanguageModel.get_peft_model(
model,
r=8, # LoRA rank - lowered from 16 to 8 to save memory
target_modules=[
"q_proj", "k_proj", "v_proj", "o_proj",
"gate_proj", "up_proj", "down_proj",
],
lora_alpha=16, # LoRA scaling factor (usually 2x rank) - lowered from 32 to 16
lora_dropout=0.05, # Supports any, but = 0 is optimized
bias="none", # Supports any, but = "none" is optimized
use_gradient_checkpointing="unsloth", # Unsloth's optimized version
random_state=3407,
use_rslora=False, # Rank stabilized LoRA
loftq_config=None, # LoftQ
)
from trl import SFTTrainer
from transformers import TrainingArguments
# Training arguments optimized for Unsloth
trainer = SFTTrainer(
model=model,
tokenizer=tokenizer,
train_dataset=train_dataset,
#eval_dataset=eval_dataset,
dataset_text_field="text",
max_seq_length=max_seq_length,
dataset_num_proc=2,
args=TrainingArguments(
per_device_train_batch_size=1,
gradient_accumulation_steps=16, # Effective batch size = 16 (8-16 recommended)
warmup_steps=20, # 2*1600/16=200, warmup ratio = 0.1, warmup steps = 23
num_train_epochs=2, # reduced from 4 to 2 to reduce training steps
learning_rate=1.5e-4, # not sure if this is the most optimal one
fp16=not torch.cuda.is_bf16_supported(),
bf16=torch.cuda.is_bf16_supported(),
logging_steps=25,
optim="adamw_8bit",
weight_decay=0.01,
lr_scheduler_type="linear",
seed=3407,
output_dir="outputs",
save_strategy="epoch",
save_total_limit=2,
dataloader_pin_memory=False,
report_to="none", # Disable Weights & Biases logging,
),
)
Fine-tuning the model
# Train the model
trainer_stats = trainer.train()
I've seen worse training losses, so the model fitting somewhat well with training data without overfitting. Should I say "not good, not terrible"?
Saving the model
import re
# Test the fine-tuned model
FastLanguageModel.for_inference(model) # Enable native 2x faster inference
messages = [{"role":"system", "content":
"""
You are an useful AI assistant. Your task is to extract clinically relevant data from electronic health record and present it in JSON-format, return nothing else but the JSON of clinical data. JSON schema is presented below:
{
"visit_datetime": "DD-MM-YYYY HH:MM:SS",
"firstname": string or null,
"lastname": string or null,
"age": integer or null,
"gender": "Male" | "Female" | "Other" | "Unknown",
"height": integer or null,
"weight": integer or null,
"bmi": float or null,
"diagnoses_old": {
"ICD10": [string],
"ICD10 description": [string]
},
"diagnoses_new": {
"ICD10": [string],
"ICD10 description": [string]
},
"medications": [
{
"ATC": string,
"Indication": string or null,
"Drug_name": string or null,
"dose": float or null,
"unit": string or null,
"form": string or null,
"note": string or null
}
],
"measurements": [
{
"Measurement": string or null,
"Value": float or null,
"Unit": string or null
}
],
"attitude": string or null,
"cognition": string or null,
"homecounty": string or null,
"smokes": "Never smoked" | "Smokes currently" | "Has quit smoking" | "No data"
}
"""
},
{"role":"user", "content": "A client arrives with a referral from specialist Annika. Person's name is Mysi Rasa. 13-05-2015 07:07:32 marked as the time of arrival. She ts somewhat malnourished. Measurements noted as height 169 cm and weight 55 kg during initial assessment. Patient doesn't smoke. Pays attention and is responsive. Displays an uplifted mood and smiles spontaneously durable the interview. Old diagnoses: R301-Dysuria. Measurements: S-EtyloE: 0.15 kU/l. S-rDer10E: 0.31 kU/l. S-L. i. Heb: 87.34 < ei-määritelty >. Medications: fesoterodine, ATC: G04BD11, indication: R301, dose: 4.0, unit: mg, form: O, tamsulosin, ATC: G04CA02, indication: R301, dose: 0.4, unit: mg, form: O. Current diagnoses: R301-Vesical tenesmus. The patient role was positioned for examination and informed of expected wait time. Reception procedures were concluded without incident."
},
]
# Explicitly set pad_token if it's not set, as it might be identical to eos_token
if tokenizer.pad_token is None:
tokenizer.pad_token = tokenizer.eos_token
# Apply chat template to get the formatted string
formatted_chat_string = tokenizer.apply_chat_template(
messages,
tokenize=False, # Get string output first
add_generation_prompt=True,
)
# Now tokenize the string to get a proper BatchEncoding object
encoded_inputs = tokenizer(
formatted_chat_string,
return_tensors="pt",
padding="max_length", # Ensure consistent padding to max_length
truncation=True, # Truncate if longer than max_length
max_length=max_seq_length, # Use the model's max_seq_length
return_attention_mask=True,
)
# Move tensors to CUDA
input_ids = encoded_inputs["input_ids"].to("cuda")
attention_mask = encoded_inputs["attention_mask"].to("cuda")
# Generate response
outputs = model.generate(
input_ids=input_ids,
attention_mask=attention_mask,
max_new_tokens=1024,
use_cache=True,
temperature=0.7,
do_sample=True,
top_p=0.9,
)
# Decode and print
response = tokenizer.batch_decode(outputs)[0]
response = response.replace('\'', '"').replace('\n', ' ').replace('nan', 'None')
tulos = re.search(r'(?<=assistant ).*?(?=\<\|im_end)', response)
print(eval(tulos.group()))
It seems that the SLM got the idea and does mine clinical data from free-text as I hoped. Now I must convert the clinical text miner model into huggingface-format and then save the model into Google Drive.
model.save_pretrained_gguf("clinical_data_miner", tokenizer, quantization_method="q4_k_m")
import os
import shutil
from google.colab import drive
drive.mount('/content/drive')
source_file = "qwen3-4b-instruct-2507.Q4_K_M.gguf"
destination_folder = "/content/drive/MyDrive/colab-downloads"
# Ensure the destination folder exists
os.makedirs(destination_folder, exist_ok=True)
destination_path = os.path.join(destination_folder, source_file)
# Move the file
shutil.move(source_file, destination_path)
print(f"Successfully moved '{source_file}' to '{destination_path}'")
Great, now that the model is saved into my Google Drive, I can download it from the to my local system. First I tried to download the clinical text miner model directly from Colab, but that was way too slow, so this was much more efficient solution.
Mining a test dataset of unstructured free-text EHR samples with fine-tuned model
Now that clinical text miner is in my local system, I can put in into production. Here I'll repurpose method presented in Mithilesh's example.
import ollama
import pandas as pd
# few dependency issues solved with:
# httpx==0.27.2
# httpcore<0.14
texts_to_process=100
schema = """
{
"visit_datetime": "DD-MM-YYYY HH:MM:SS",
"firstname": string or null,
"lastname": string or null,
"age": integer or null,
"gender": "Male" | "Female" | "Other" | "Unknown",
"height": integer or null,
"weight": integer or null,
"bmi": float or null,
"diagnoses_old": {
"ICD10": [string],
"ICD10 description": [string]
},
"diagnoses_new": {
"ICD10": [string],
"ICD10 description": [string]
},
"medications": [
{
"ATC": string,
"Indication": string or null,
"Drug_name": string or null,
"dose": float or null,
"unit": string or null,
"form": string or null,
"note": string or null
}
],
"measurements": [
{
"Measurement": string or null,
"Value": float or null,
"Unit": string or null
}
],
"attitude": string or null,
"cognition": string or null,
"homecounty": string or null,
"smokes": "Never smoked" | "Smokes currently" | "Has quit smoking" | "No data"
}
"""
# Initialize the Ollama client
client = ollama.Client()
def write_text_to_file(file_path, text):
#Write text to a file.
with open(file_path, "w", encoding="utf-8") as file:
file.write(text)
def mine_text(client, model, text):
mine_prompt = f"""You are an useful AI assistant. Your task is to extract clinically relevant data from electronic health record and present it in JSON-format, return nothing else but the JSON of clinical data. Use only quotations (") for wrapping str values. JSON schema is presented below:
{schema}
Extract clinical contents of the following text into JSON-format as presented above: {text}"""
response = client.generate(model=model, prompt=mine_prompt)
response = response.get("response", "")
return response
# Define the path to the input and output text files
base_path = "/clinical_text_miner/test_directory/"
# make directory for output
output_path = "miner_output/"
if os.path.exists(output_path) != True:
os.mkdir(output_path)
# Read the text from the file
#text = read_text_from_file(input_file_path)
df = pd.read_excel(base_path+ "synthetic_patient_texts_for_testing.xlsx")
df = df.loc[0:texts_to_process-1] # we'll test just first 100 rows
# Mine text
print(f"Iterating through {len(df)} texts.")
for i in range(len(df)):
text = mine_text(client, "clinical_text_miner", df["text corrupter"][i])
# Write the anonymized text to the output file
write_text_to_file(output_path + df["id"][i] + ".txt", text)
print(f"Text {i} processed and saved to {output_path}{df["id"][i]}.txt.")
# Print a confirmation message
print(f"Mined texts have been written and stored into to the {output_path[:-1]} directory.")
That took a good while. About six and a half hours to exact. with only CPU resources the AI utilization drags down to a slog. Since my local system only has CPU-resources, the processing time takes exponentially longer compared to GPU.
Formatting fine-tuned model's output
I'll iterate over the processed files and store them into dftest dataframe.
import os
import pandas as pd
output_path = "miner_output/"
dftest = pd.DataFrame({"id":[], "json_raw": []})
directory = os.fsencode(output_path)
for file in os.listdir(directory):
filename = os.fsdecode(file)
if filename.endswith(".txt"):
with open(output_path+filename) as f: s = f.read()
dftest = pd.concat([dftest, pd.DataFrame({"id":[filename[:-4]], "json_raw": [s]})], ignore_index=True)
else:
continue
I checked the contents of the file and clearly the mining process wasn't completely perfect. The main problems are caused by how Pandas handles missing (null) values and texts containing both quotation marks (") and apostrophes (') messing with transforming the string into dictionary. These artefacts cause problems with eval()-function so I first must clean the texts before I can pass them into json.dumps(), which will properly turn the texts, turned into dictionary datatype, into JSON files
import re
# this will be a preliminary step where I first transform dftest's string values into dictionaries
prejsons = []
for i in range(len(dftest)):
try:
tmp = dftest.json_raw.str.replace('\n', '', regex=True).str.replace(r"\'", '"', regex=True)\
.str.replace('nan', 'None', regex=True).str.replace('null', 'None', regex=True)\
.str.replace('(?<=[a-zA-Z])(?:\")(?=[a-zA-Z])', "'", regex=True)[i]
prejsons.append(eval(tmp))
except:
try:
# if the key has missing quotes, I'll character sequence next to colon and wrap it in quotes
s=re.search(r'([^\s]+)[a-öA-Ö](?:: .)', tmp).start()
e=re.search(r'([^\s]+)[a-öA-Ö](?:: .)', tmp).end()-3
tmp2 = tmp[:e] + '"' + tmp[e:] # ending first so positional index is not messed up
tmp2 = tmp2[:s] + '"' + tmp2[s:]
prejsons.append(eval(tmp2))
except:
# if that doesn't work either, then maybe the error lies in corrupted numbers
# first we'll find all unquoted values and for each, we'll try to read it as some datatype
# if it produces value error, then we have found our match
for m in re.findall(r'"(\w+)": (\w+)', tmp):
try:
eval(m[1]) # eval test the match, if it doesn't work, we'll change it to string
except:
# print the value that is to be turned into string
print(f'Value error in sample {i}: "{m[0]}": {m[1]} -> {m[0]}: "{m[1]}"')
# turn the value into string
tmp2 = tmp.replace(str('"' + m[0] + '": ' + m[1]), str('"' + m[0] + '": ' + '"' + m[1]+ '"'))
prejsons.append(eval(tmp2))
One file in sample 48 couldn't initially be formatted into a dictionary, because text corrupter added a string character into an age value that was supposed to be a integer. Despite that, everything else seems to be in order. Let's check the EHR free-text data at that index.
base_path = "/clinical_text_miner/test_directory/"
df = pd.read_excel(base_path+ "synthetic_patient_texts_for_testing.xlsx")
df = df.loc[0:99] # we'll test just first 100 rows
print(df[df['id']==dftest['id'][48]]['text corrupter'].values)
And there you have it. The model acted as expected and dutifully extracted age information from unstructured EHR into the age field that was designated as integer datatype in the AI prompt. This lead to a string value not being wrapped in quotes that ended up in error with eval-funtion. In the future in designing text extraction prompts, I may have to designate all fields as string datatype, which should be interpreted again into proper datatype after extraction result is interpreted as a dictionary.
Now, let's convert these dictionaries into JSON-format.
import json
# here I'll turn dictionaries into JSON
# I'll still use iterating over range(len()) since I can follow errors in process via index
jsons = []
for i in range(len(prejsons)):
try:
jsons.append(json.dumps(prejsons[i],indent=4))
except:
print(f"error on index {i}")
jsons.append(None)
Let's see how it looks.
print(jsons[48])
Looks promising! Despite the unicode corruption for umlaut characters, like in homecounty value, the results are quite impressive. This demonstrates that it is technically possible to extract several clinically relevant variables from free-text EHR data and store it in structued format. Next step would be to actually test my model's performance with our test dataset against ground truth and see how well text mining has performed.
Testing fine-tuned model's output
I'll do testing the lazy way and compare mined json to ground truth json with SequenceMatcher library, but first I'll prepare the testing dataframe.
groundtruth = []
for i in range(len(df)):
try:
tmp = df.json.str.replace('\n', '', regex=True).str.replace(r"\'", '"', regex=True)\
.str.replace('nan', 'None', regex=True).str.replace('null', 'None', regex=True)\
.str.replace('(?<=[a-zA-Z])(?:\")(?=[a-zA-Z])', "'", regex=True)[i]
groundtruth.append(eval(tmp))
except:
print(f"error at index: {i}")
groundtruth.append(tmp)
# instead of comparing raw json, let's compare dictionaries instead since they are easier to handle
df['json_test'] = groundtruth # save jsons-turned-dictionaries for testing
dftest['json_pred'] = prejsons # prejsons are actually dictionaries so we can use them
dftest = df[['id', 'json_test']].merge(dftest[['id','json_pred']], on='id')
from difflib import SequenceMatcher
def similar(a, b):
# I'll set the strings to lowercase just to be sure
return SequenceMatcher(None, str(a).lower(), str(b).lower()).ratio()
result = pd.DataFrame({'visit_datetime': [],'firstname': [],'lastname': [],'age': [],
'gender': [],'height': [],'weight': [],'bmi': [],'diagnoses_old': [],
'diagnoses_new': [],'medications': [],'measurements': [],
'attitude': [],'cognition': [],'homecounty': [],'smokes': []})
for i in range(len(dftest)):
scores={}
for key in dftest.json_test[i].keys():
score=similar(dftest.json_test[i][key], dftest.json_pred[i][key])
scores[key] = [score]
result = pd.concat([result,pd.DataFrame(scores)], ignore_index=True)
for key in result.columns:
print(f"{key}: {round(result[key].mean(),4)}")
And here are the results comparing text similarity ratios. Lastname, gender, height and homecounty information were mined perfectly. Visitation datetime, firstname, age, weight and measurement information were mined almost perfectly. Diagnosis and medication information was mined decently, but smoking status, attitude, cognition and especially bmi were lacking. I presume some of the impact against ratio-score for variables with mining ratio-score above 0.95 was due to text corruption, but for those under that treshold is not easy to rationalize with just natural consequence of added data noise.
Next, I'll calculate how well the model mined ICD10 diagnoses with precision, recall and F1-scoring.
# result in long table form
result_l = pd.DataFrame({'Key': [], 'Type': [], 'Accuracy' : [], 'Precision': [], 'Recall': [], 'F1-Score': []})
def getf1(key, t, list1, list2):
set1=set(list1)
set2=set(list2)
# prediction is true if both sets lack values
if len(set1)==0 and len(set2)==0:
return {'Key': [key], 'Type': t, 'Accuracy' : [True], 'Precision': [True], 'Recall': [True], 'F1-Score': [True]}
# prediction is false if both sets lack values
elif len(set1)==0 or len(set2)==0:
return {'Key': [key], 'Type': t, 'Accuracy' : [False], 'Precision': [False], 'Recall': [False], 'F1-Score': [False]}
else:
true_positives = [item for item in set1 if item in set2] # items in both sets, true positive
false_negatives = [item for item in set1 if item not in set2] # items in set1 and not in set2, false negative
false_positives = [item for item in set2 if item not in set1] # items in set2 and not in set1, false positive
acc = len(true_positives)/(len(true_positives)+len(false_negatives)+len(false_positives))
pre = len(true_positives)/(len(true_positives)+len(false_positives))
rec = len(true_positives)/(len(true_positives)+len(false_negatives))
# have to use try and except if true positives end up being 0
try:
f1 = round(2*((pre*rec)/(pre+rec)),2)
except:
return {'Key': [key], 'Type': t, 'Accuracy' : [acc], 'Precision': [pre], 'Recall': [rec], 'F1-Score': [0]}
return {'Key': [key], 'Type': t, 'Accuracy' : [acc], 'Precision': [pre], 'Recall': [rec], 'F1-Score': [f1]}
for i in range(len(dftest)):
for key in ['diagnoses_old', 'diagnoses_new']:
score=getf1(key, 'code', dftest.json_test[i][key]['ICD10'], dftest.json_pred[i][key]['ICD10'])
result_l = pd.concat([result_l,pd.DataFrame(score)], ignore_index=True)
for i in range(len(dftest)):
for key in ['diagnoses_old', 'diagnoses_new']:
score=getf1(key, 'description', dftest.json_test[i][key]['ICD10 description'], dftest.json_pred[i][key]['ICD10 description'])
result_l = pd.concat([result_l,pd.DataFrame(score)], ignore_index=True)
# Print each analyzed key
for key in result_l.Key.unique():
for t in result_l.Type.unique():
for column in result_l.columns[2:]:
print(f"{key} {t} - {column}: {round(result_l[(result_l.Key==key) & (result_l.Type==t)][column].mean(),4)}")
Here we can see that the model clearly performs better with short and more comprehensible ICD10 codes but falters with diagnosis code descriptions. Text similarity ratio-scoring is more forgiving, since it calculates similarity of each character in fields under comparison, whereas in true positive/negative comparison even slightest variation counts as unique value and as either complete match or mismatch. There is less of a chance to make a mistake with short codes, but performance score receives a heavy penalty even for the slightest mistake in long descriptions. Here I may have made a mistake since the model in some cases interprets that description ought to contain only ICD10 description but the ground truth contains both code and description in the ICD10 description field. By either removing codes from description field or by naming "ICD10 description" to "ICD10 code and description" would've perhaps mitigated this problem.
Comparing fine-tuned model's performance against non-fine-tuned model
I think this will suffice in providing a general overview on model's mining performance. However, this explains only my fine-tuned model's performance but that doesn't explain much for the efficacy of the fine-tuning itself. Therefore I have to compare it to the original Qwen3-4b-Instruct-2507 model. Luckily I don't have transform another Gwen3 to GGUF, since unsloth has already released one and I can easily pull it from huggingface with following terminal command:
ollama pull https://huggingface.co/unsloth/Qwen3-4B-Instruct-2507-GGUF
Now I will run all previous steps again but with non-fine-tuned Qwen3 and assess it against my fine-tuned model.
import ollama
import pandas as pd
# few dependency issues solved with:
# httpx==0.27.2
# httpcore<0.14
texts_to_process=100
# Initialize the Ollama client
client = ollama.Client()
def write_text_to_file(file_path, text):
#Write text to a file.
with open(file_path, "w", encoding="utf-8") as file:
file.write(text)
def mine_text(client, model, text):
# use the same schema as with fine-tuned model
mine_prompt = f"""You are an useful AI assistant. Your task is to extract clinically relevant data from electronic health record and present it in JSON-format, return nothing else but the JSON of clinical data. Use only quotations (") for wrapping str values. JSON schema is presented below:
{schema}
Extract clinical contents of the following text into JSON-format as presented above: {text}"""
response = client.generate(model=model, prompt=mine_prompt)
response = response.get("response", "")
return response
# Define the path to the input and output text files
base_path = "/clinical_text_miner/test_directory/"
#input_file_path = base_path + "data_for_input.txt"
#output_file_path = base_path + "data_for_output.txt"
# make directory for output
output_path = "Qwen3_output/"
if os.path.exists(output_path) != True:
os.mkdir(output_path)
# Read the text from the file
#text = read_text_from_file(input_file_path)
df = pd.read_excel(base_path+ "synthetic_patient_texts_for_testing.xlsx")
df = df.loc[0:texts_to_process-1] # we'll test just first 100 rows
# Mine text
print(f"Iterating through {len(df)} texts.")
for i in range(len(df)):
text = mine_text(client, "huggingface.co/unsloth/Qwen3-4B-Instruct-2507-GGUF:latest", df["text corrupter"][i])
# Write the anonymized text to the output file
write_text_to_file(output_path + df["id"][i] + ".txt", text)
print(f"Text {i} processed and saved to {output_path}{df["id"][i]}.txt.")
#write_text_to_file(output_file_path, mined_text)
#df_final = pd.DataFrame({"id":df["id"], "truth": df["json"], "pred": mined_texts})
#df_final.to_excel('mined_texts.xlsx')
# Print a confirmation message
print(f"Mined texts have been written and stored into to the {output_path[:-1]} directory.")
import os
import pandas as pd
# let's name our temporary dataframe as dfq as in "dataframe-Qwen"
dfq = pd.DataFrame({"id":[], "json_raw": []})
output_path = "Qwen3_output/"
directory = os.fsencode(output_path)
for file in os.listdir(directory):
filename = os.fsdecode(file)
if filename.endswith(".txt"):
with open(output_path+filename) as f: s = f.read()
dfq = pd.concat([dfq, pd.DataFrame({"id":[filename[:-4]], "json_raw": [s]})], ignore_index=True)
else:
continue
# again, first I'll transform dfq's string values into dictionaries
prejsons = []
for i in range(len(dfq)):
try:
tmp = dfq.json_raw.str.replace('\n', '', regex=True).str.replace(r"\'", '"', regex=True)\
.str.replace('nan', 'None', regex=True).str.replace('null', 'None', regex=True)\
.str.replace('(?<=[a-zA-Z])(?:\")(?=[a-zA-Z])', "'", regex=True)[i]
prejsons.append(eval(tmp))
except:
try:
# if the key has missing quotes, I'll character sequence next to colon and wrap it in quotes
s=re.search(r'([^\s]+)[a-öA-Ö](?:: .)', tmp).start()
e=re.search(r'([^\s]+)[a-öA-Ö](?:: .)', tmp).end()-3
tmp2 = tmp[:e] + '"' + tmp[e:] # ending first so positional index is not messed up
tmp2 = tmp2[:s] + '"' + tmp2[s:]
prejsons.append(eval(tmp2))
except:
# if that doesn't work either, then maybe the error lies in corrupted numbers
# first we'll find all unquoted values and for each, we'll try to read it as some datatype
# if it produces value error, then we have found our match
for m in re.findall(r'"(\w+)": (\w+)', tmp):
try:
eval(m[1]) # eval test the match, if it doesn't work, we'll change it to string
except:
# print the value that is to be turned into string
print(f'Value error in sample {i}: "{m[0]}": {m[1]} -> {m[0]}: "{m[1]}"')
# turn the value into string
tmp2 = tmp.replace(str('"' + m[0] + '": ' + m[1]), str('"' + m[0] + '": ' + '"' + m[1]+ '"'))
prejsons.append(eval(tmp2))
# rest of samples that cannot be fixed are stored in marked and stored in prejsons list
#print(f"error on index {i}")
#prejsons.append(tmp)
# save the results in dataframe
dfq['json_q_pred'] = prejsons
dftest = dftest.merge(dfq[['id','json_q_pred']], on='id')
# turn dictionaries into JSON
result_q = pd.DataFrame({'visit_datetime': [],'firstname': [],'lastname': [],'age': [],
'gender': [],'height': [],'weight': [],'bmi': [],'diagnoses_old': [],
'diagnoses_new': [],'medications': [],'measurements': [],
'attitude': [],'cognition': [],'homecounty': [],'smokes': []})
for i in range(len(dftest)):
scores={}
for key in dftest.json_test[i].keys():
score=similar(dftest.json_test[i][key], dftest.json_q_pred[i][key])
scores[key] = [score]
result_q = pd.concat([result_q,pd.DataFrame(scores)], ignore_index=True)
I have used Qwen3 to mine clinical texts and stored them into dictionaries and JSON-format. Now it is possible to compare my fine-tuned model's performance against non-fine-tuned model. I'll format fine-tuned and non-fine-tuned models' results in long form so they can be plotted in a bar chart.
result['set']='Fine-tuned'
result_q['set']='Not fine-tuned'
result_m = pd.melt(result, id_vars=['set'], value_vars=['visit_datetime', 'firstname',
'lastname', 'age', 'gender',
'height','weight', 'bmi',
'diagnoses_old', 'diagnoses_new',
'medications', 'measurements',
'attitude', 'cognition',
'homecounty', 'smokes'])
result_q['set']='Not fine-tuned'
result_q_m = pd.melt(result_q, id_vars=['set'], value_vars=['visit_datetime', 'firstname',
'lastname', 'age', 'gender',
'height','weight', 'bmi',
'diagnoses_old', 'diagnoses_new',
'medications', 'measurements',
'attitude', 'cognition',
'homecounty', 'smokes'])
results_all = pd.concat([result_m, result_q_m])
The moment of dubious truth!
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(rc={'figure.figsize':(10,4)})
p=sns.barplot(results_all, x='variable', y='value', hue='set', errorbar=None)
p.set_xticklabels(labels=results_all['variable'].unique(),rotation=45, ha='right')
sns.move_legend(p, "upper left", bbox_to_anchor=(1, 1))
p.set_title('Text similarity ratio-scores between fine-tuned and not fine-tuned models')
Now this is interesting. By using text similarity ratio to assess the performance, it is mostly similar between models, with fine-tuned model slightly outperforming non-fine-tuned model in many clinical variables except for smoking status where non-fine-tuned model takes the lead. Let's evaluate performance with F1-score.
from sklearn.metrics import f1_score
import warnings
warnings.simplefilter('ignore')
dftest_truth = pd.DataFrame(dftest['json_test'].to_list()) # true values
dftest_pred = pd.DataFrame(dftest['json_pred'].to_list()) # mined values
dftest_pred_q = pd.DataFrame(dftest['json_q_pred'].to_list()) # true values
result_f1 = pd.DataFrame({'Variable':[],'Set':[],'F1-score':[]})
# again, I'll omit diagnoses, measures and medications, since they are nested lists and dicts
for key in ['visit_datetime', 'firstname', 'lastname', 'age', 'gender', 'height',
'weight', 'bmi', 'attitude', 'cognition', 'homecounty', 'smokes']:
f1_tmp1 = f1_score(dftest_truth[key].astype(str), dftest_pred[key].astype(str), average='weighted')
result_f1 = pd.concat([result_f1, pd.DataFrame({'Variable':[key],'Set':['Fine-tuned'],'F1-score':[f1_tmp1]})])
f1_tmp2 = f1_score(dftest_truth[key].astype(str), dftest_pred_q[key].astype(str), average='weighted')
result_f1 = pd.concat([result_f1, pd.DataFrame({'Variable':[key],'Set':['Not fine-tuned'],'F1-score':[f1_tmp2]})])
sns.set_theme(rc={'figure.figsize':(8,4)})
p=sns.barplot(result_f1, x='Variable', y='F1-score', hue='Set', errorbar=None)
p.set_xticklabels(labels=result_f1['Variable'].unique(),rotation=45, ha='right')
sns.move_legend(p, "upper left", bbox_to_anchor=(1, 1))
p.set_title('F1-scores between fine-tuned and not fine-tuned models')
F1-scoring shows similar results, but with greater dispersion between 1 and 0, due to F1-scoring punishing more severely mislabeling.
I'll also compare ICD10 code and description mining performance with F1-scoring.
result_q_l = pd.DataFrame({'Key': [], 'Type': [], 'Accuracy' : [], 'Precision': [], 'Recall': [], 'F1-Score': []})
for i in range(len(dftest)):
for key in ['diagnoses_old', 'diagnoses_new']:
score=getf1(key, 'code', dftest.json_test[i][key]['ICD10'], dftest.json_q_pred[i][key]['ICD10'])
result_q_l = pd.concat([result_q_l,pd.DataFrame(score)], ignore_index=True)
for i in range(len(dftest)):
for key in ['diagnoses_old', 'diagnoses_new']:
score=getf1(key, 'description', dftest.json_test[i][key]['ICD10 description'], dftest.json_q_pred[i][key]['ICD10 description'])
result_q_l = pd.concat([result_q_l,pd.DataFrame(score)], ignore_index=True)
result_q_l['Dataset'] = 'Not fine-tuned'
result_l['Dataset'] = 'Fine-tuned'
result_l = pd.concat([result_l, result_q_l])
result_l[(result_l.Key=='diagnoses_old') & (result_l.Type=='code')]
sns.set_theme(rc={'figure.figsize':(4,4)})
p=sns.barplot(result_l[result_l.Key=='diagnoses_old'], x='Type', y='F1-Score', hue='Dataset', errorbar=None, estimator='mean')
#p.set_xticklabels(labels=result_f1['Variable'].unique(),rotation=45, ha='right')
sns.move_legend(p, "upper left", bbox_to_anchor=(1, 1))
p.set_title('F1-scores in mining performance of old diagnosis code and description')
p=sns.barplot(result_l[result_l.Key=='diagnoses_new'], x='Type', y='F1-Score', hue='Dataset', errorbar=None, estimator='mean')
#p.set_xticklabels(labels=result_f1['Variable'].unique(),rotation=45, ha='right')
sns.move_legend(p, "upper left", bbox_to_anchor=(1, 1))
p.set_title('F1-scores in mining performance of new diagnosis codes and descriptions')
And there you have it. As text similarity ratio is more forgiving compared to complete similarity comparison, fine-tuned model takes the prize in almost all fields except for smoking status. The discrepancy between smoking status prediction compared to other variables may be due to a small training dataset, but I doubt this is the case. I reckon more likely cause is discrepancy between input texts and output labels and lack of diversity in smoking status labels, that may have had confounding effect on the model.
After having looked more in depth in all 17 cases where fine-tuned model had predicted falsely whereas non-fine-tuned model predicted correctly, it was clear that the problem was with overfitting. Fine-tuned model put too much reliance on the label "has quit smoking", putting a labeling emphasis on the word "smoked", usually ignoring important context around the term related to smoking. "Never smoked" -declarations in text were many times interpreted as "has quit smoking" and few "no data" -statuses were interpreted as never smoked. This problem could have been mitigated in the training phase by using validation dataset and more training steps with smaller learning rate, but I couldn't do so due to memory constraints.
Discussion
In this blog post I have fine-tuned a small Qwen3-4B-Instruct-2507 language model to mine clinically relevant text from unstructured free-text EHR data and output it in structured JSON-format. I evaluated model's mining performance with text similarity ratio-scoring and F1-scoring, where success was moderate for datetime, name, gender, height, weight, homecounty and diagnosis values. prediction of attitude, smoking status and cognition were less than moderate and bmi prediction was poor. Fine-tuning provided from remarkable to minor improvements in data mining, but smoking status mining decreased in performance. This method presents not only the possibilities but also several challenges in fine-tuning, many of which are already familiar in machine learning training, such as dataset collection and curation, setting training hyperparameters, and overfitting. Also, and in my case most significantly, fine-tuning can be limited by memory constraints, which necessitates setting suboptimal training parameters, decreasing the performance of fine-tuned model.
Sources
- fine-tuning LLMs instructions on how to fine-tune a model
- Google Colab free fine-tuning GPU resources
- Enhancing text anonymization with Ollama instructions on how to implement a fine-tuned model in production
- Qwen3-4B-Instruct-2507 a model that was used for fine-tuning
- Qwen3-4B-Instruct-2507-GGUF a comparison model that was Ollama compatible (GGUF)
- Unsloth for speeding up fine-tuning process

